Jumping genes
Published in Ecology & Evolution
Bacterial genomes are remarkably changeable, adapting to new environments by deleting genes, and acquiring different ones from their neighbours. In one metaphor, bacteria are like smart phones and the genes they swap like apps (Young, 2016). But rather than being downloaded, genes transfer between bacteria largely thanks to plasmids — genetic elements that can copy themselves, as well as any genetic cargo, between neighbouring bacteria. Technology of the 1980s provides a more suitable (though maybe less familiar) metaphor: if bacteria are computers, the genes they swap are programs, and plasmids are like floppy disks (they also tend to have a hole in the middle!).

Much of what we know about plasmids is based on short experiments under defined laboratory conditions, or on the painstaking work of bioinformaticians sifting through genomes from clinical or environmental samples. At the Brockhurst lab in the University of Sheffield we use a complementary approach to explore plasmids: experimental evolution. By setting up replicate bacterial populations in the lab, under different conditions, and culturing them for hundreds of generations, we can tease apart the conditions favouring plasmid maintenance and spread.

In this study we wanted to complicate things by introducing an additional bacterial species. Moreover, we wanted our experiments to better represent conditions faced by bacteria ‘in the wild’. Inspired by Angus Buckling’s group at the University of Exeter, we began experimenting with soil microcosms. These consist of a small volume of autoclaved potting soil in a glass vial, which is mixed and refreshed every few days. By adding small amounts of mercury, we could select for bacteria carrying a mercury resistance plasmid, by killing or inhibiting their plasmid-free competitors. While this set-up cannot capture the complexity of life outside the lab, it goes beyond rich liquid broth by providing a three-dimensional growth matrix and complex food source. It is also reasonably compact, allowing scores of independent populations to be cultured in parallel, and relatively consistent: results were generally no more variable than those in broth (Hall et al., 2015).

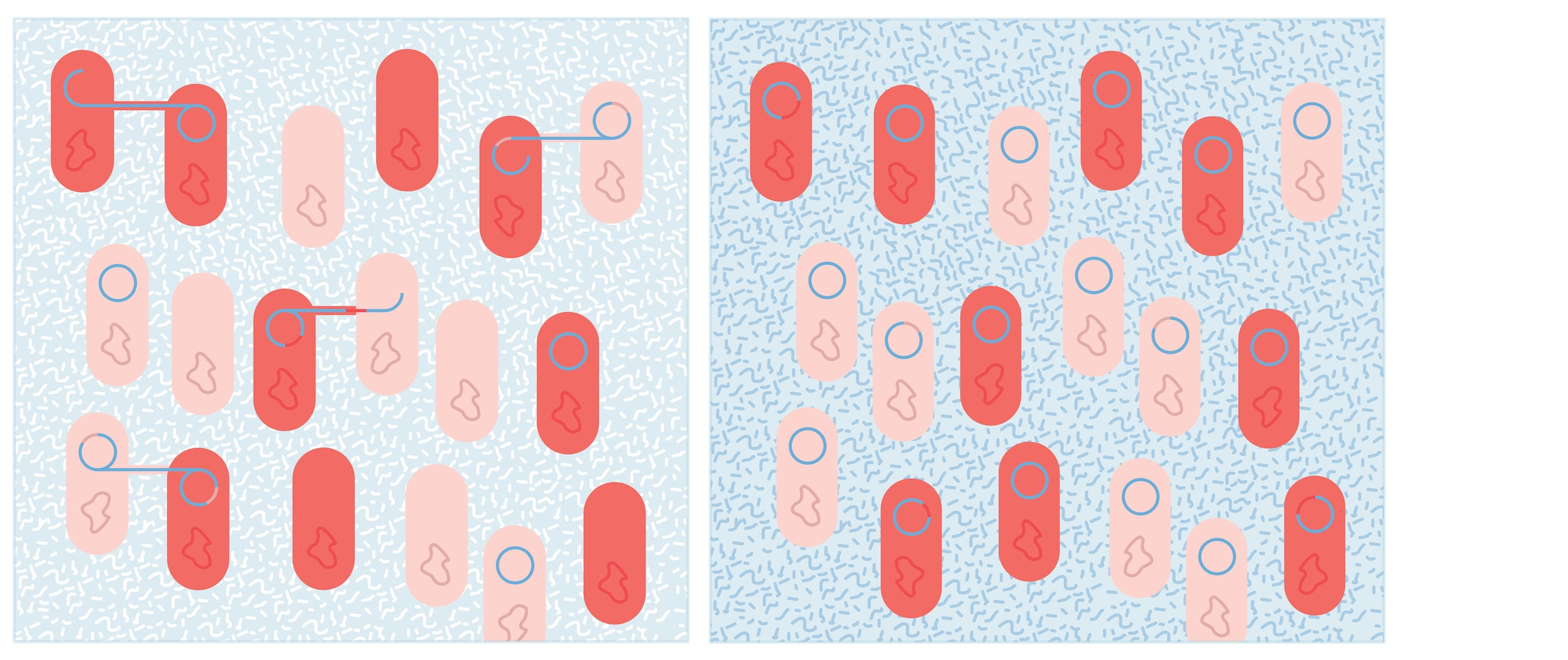
Like an entry to a flower show (an alternative destiny of our bag of soil), our microcosms required careful tending over a long period. Nine months of close attention (and over 25,000 PCR reactions) revealed that when grown separately, our bacterial species differed in their ability to maintain the plasmid, but when cultured together, the plasmid could be better maintained across the community (Hall et al., 2016).
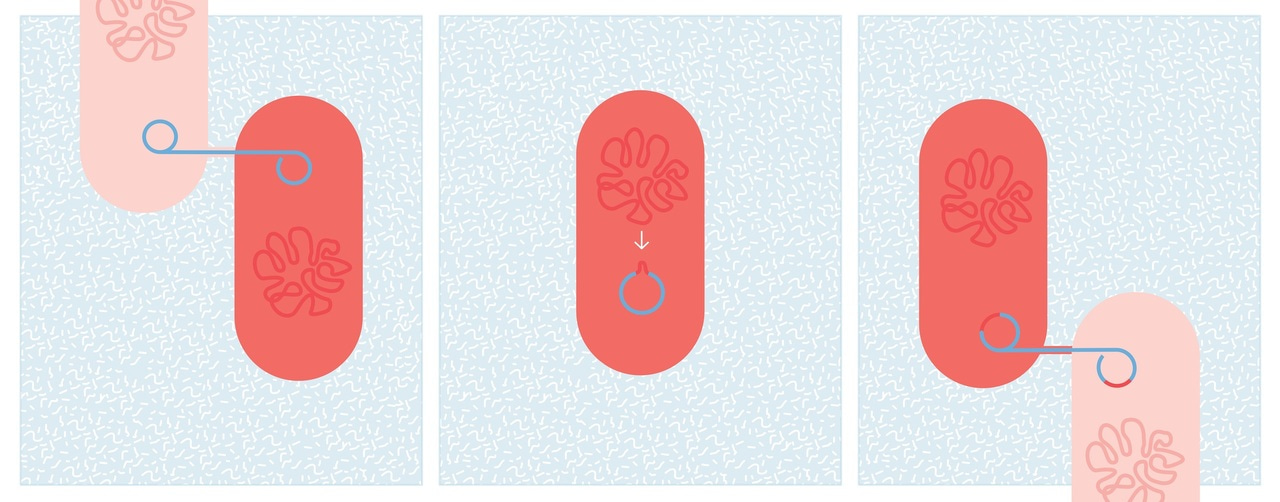
We were curious to see how this dynamic affected the evolution of bacteria and plasmid, so we sequenced clones from the beginning and the end of the experiment. Early in the analysis we had a suggestion that something interesting had happened: coverage plots showed clones from one species with part of the genome of the other, and vice-versa. The full results took us some time to wrap our heads around, as most sequence analysis pipelines are geared towards looking for small changes. But they confirmed what we had seen initially: a lot of gene movement, both within chromosome and between plasmid and chromosome. Most strikingly, a region of one species’ chromosome had been picked up by the plasmid, and transferred into the other species — a pattern found repeated in several populations. Microbial genomes are clearly highly dynamic, and genes can readily hop on and off plasmids to new hosts.
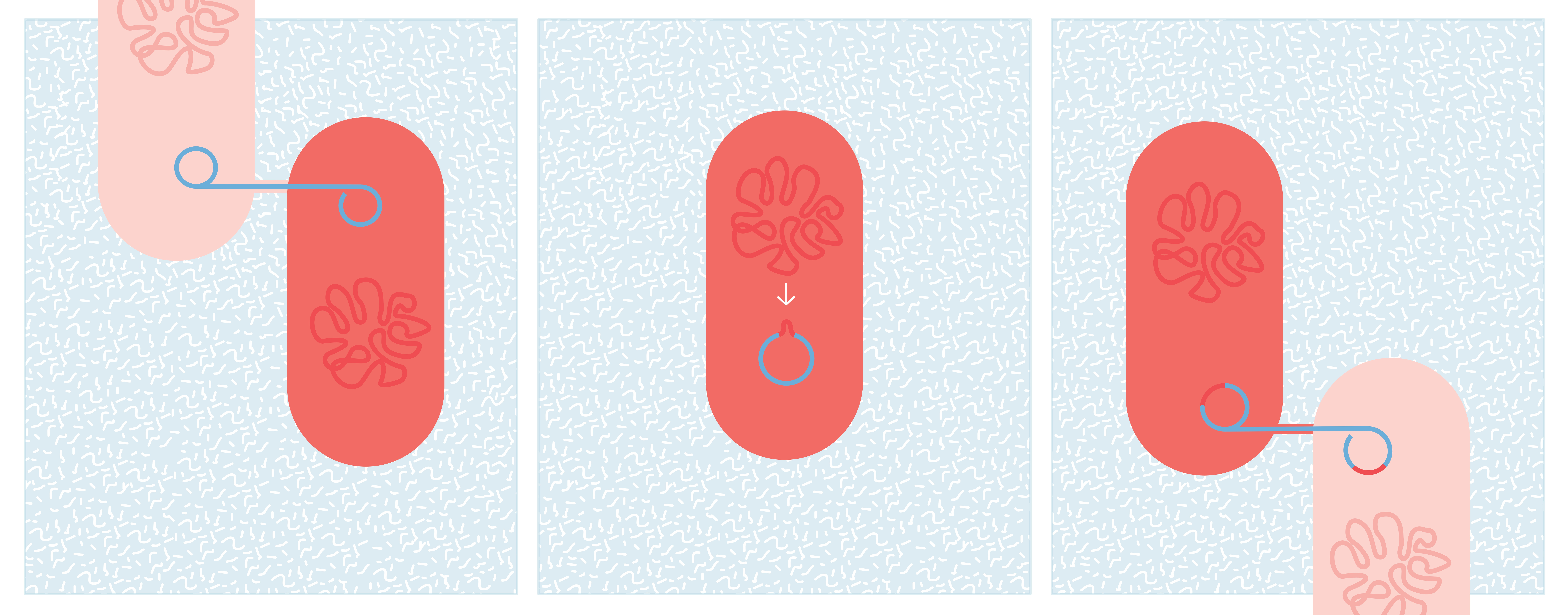
We were intrigued that the plasmid acted as a shuttle for gene exchange only where it was not being selected for (i.e. no mercury added). This seemed strange: surely a plasmid would be better at picking up and spreading genes to new hosts when it was beneficial? However, just as a disk drive can only hold one disk, mechanisms such as surface exclusion and incompatibility mean that bacteria that are already carrying a resistance plasmid cannot easily receive another copy. Selection for plasmid carriage results in more plasmid-carrying bacteria, so there are fewer potential recipients of gene exchange. In short, where a plasmid is beneficial, bacteria are more likely to receive it ‘vertically’ from their parent than ‘horizontally’ from their neighbours, restricting its activity as a gene shuttle.
It’s been very interesting discussing this data with scientists who study gene transfer ‘in the wild’. There’s increasing concern that places like hospital plumbing, waste-water treatment plants, or animal manure, are sites of resistance gene exchange. While these environments can and do become contaminated with antibiotics, they may also be locations where selection is relaxed, providing opportunities for plasmids to move other genes between species. We hope that our results stimulate further work to explore this possibility.
The paper in Nature Ecology & Evolution is here: http://go.nature.com/2vXuYhy




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in