Manually annotated and curated dataset of diverse weed species in maize and sorghum for computer vision
Published in Sustainability, Agricultural & Food Science, and Statistics

Background
Weeds are unwanted plants that can affect crop growth by competing for resources such as nutrients and sunlight. Weeds can also harbor insects and diseases, creating additional challenges. The crop is not able to grow at its optimum, resulting in reduced productivity. Farmers try to mitigate these losses by removing weeds, which increases labor and costs. A common practice is to spray herbicides over the entire field, which not only decimates the weeds, but also reacts with the crop and soil. This can be harmful to the biota and the surrounding environment. Sustainable weed management is essential to ensure food production while preserving ecosystems. In organic farming, for example, the use of herbicides is limited due to environmental concerns, resulting in the need for non-chemical and site-specific weed management strategies. This increases the workload and costs for farmers by requiring more sophisticated machinery. A major milestone towards a more automated and site-specific weed management is the accurate and efficient sensing and mapping of weeds and crops, which is challenging due to their phenotypic similarity. Therefore, computer vision-based models could be used as a tool to automatically extract relevant features from the images to better distinguish weeds from crops, but these models typically require large, high-quality data sets.
Data Generation
With the Moving Fields Weed Dataset (MFWD), we aim to support the development of deep learning technologies for precision agriculture and develop strategies for sustainable weed management. We focused on a wide variety of weed species and collected a large number of images in a high-tech facility, capturing the growth dynamics of 28 weed species commonly found in maize and sorghum fields in Germany (see examples in Figure 1).

Example of each plant species with corresponding EPPO code.
Growing weeds on purpose
Considerable effort went into selecting different weed species to capture the wide variety of weeds found in fields in Germany. We grew the weeds in a controlled environment, sown in microplots (40 × 30 × 22 cm external dimensions) using a high-throughput phenotyping system. This system was capable of automatically watering, weighing, and photographing the plants. To our surprise, some weed species were difficult to grow. The germination rate was very low and we experimented with different vernalization techniques to break the seed dormancy and achieve a higher germination rate.
The experiment covered the growth period from sowing to harvest and captured the changes in plant appearance over time. The imaging process involved the use of an imaging cabin equipped with cameras and fluorescent lamps to take high-resolution top-down images of the microplots (see Figure 2a). Plants were photographed frequently - at least daily - and tracked over time to observe the growth dynamics of each individual plant. Some plants were also removed during growth if they would overlap heavily with other plants. This decision made the annotation step more feasible, especially for weeds with a high germination rate. In total, more than 94,000 images were taken during the period from 16.06.2021 to 13.12.2021.
Annotation Process
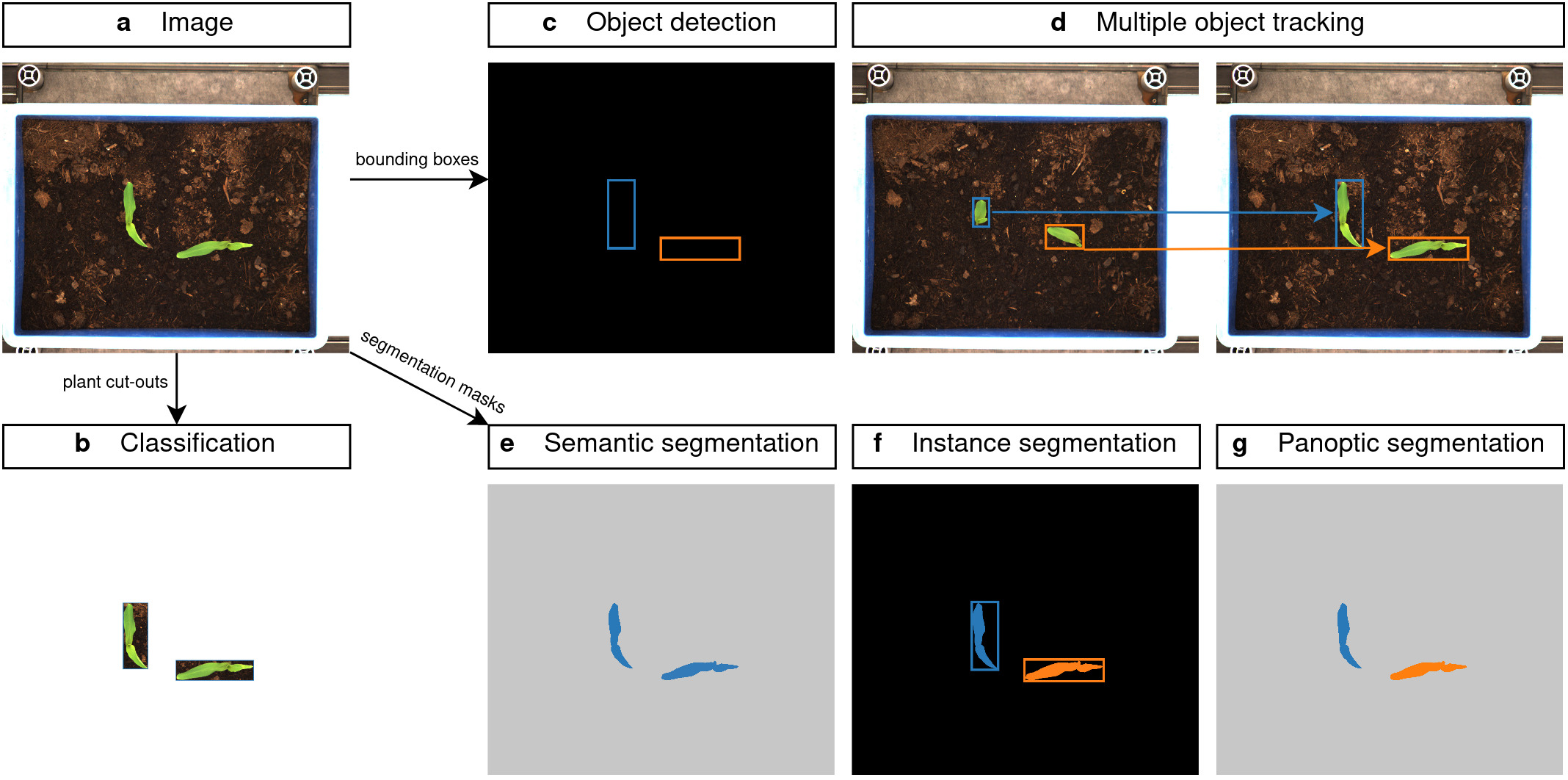
We used the open source software CVAT (Computer Vision Annotation Tool) to manually annotate the dataset. Unlike crops, the seeds of some weed species were rarely pure, so additional weed species germinated during the experiments that we did not plant. We therefore used the generic term "weed" for these unknown weed species and had them identified by an expert at a later stage. This resulted in over 200,000 bounding box annotations of the plants, which can be used as plant cut-outs for classification tasks (Figure 2b) or in object detection tasks (Figure 2c). CVAT provided a simple interface for tracking multiple objects in an image set, which allowed us to add time-series growth information for each individual plant (see Figure 2d).
Bounding box information may not be precise enough for some weed removal tasks, so we enriched our dataset with semantic and instance-level segmentation labels (see Figure 2 e-g). Therefore, we used another open source software called GIMP (GNU Image Manipulation Program) to manually paint each pixel of the plants. We selected 14 different weed species and labeled one microplot each, so that we could capture the entire growth of each plant. In total, we ended up with > 2,000 instance segmentation masks of individual plant samples. Some weed species could not be pixel annotated due to the high complexity of the plants.
Dataset
The dataset is deposited at the Digital Library of the Technical University of Munich (https://mediatum.ub.tum.de/1717366) and has a total size of > 900 GB. The image data is stored in PNG format to ensure high quality without compression artifacts. We have also released the compressed JPEG images so that researchers with less bandwidth or storage space can use our dataset. The bounding box and tracking information for all annotations is stored in a CSV file named "gt.csv". Additionally, there are folders for the segmentation and instance-level masks.
Using the dataset
There is a wide range in germination success between species, so some microplots contained only one growing plant, and others had more than 100 germinated plants. In cases of extreme growth rates, we annotated only those plants that grew for a longer period of time without being removed due to overlap, but in general, each species has at least a few microplots that were fully annotated with bounding boxes and can be used for supervised object detection. Also, microplots with partially annotated bounding boxes could be an additional source in self-supervised pre-training or in a weakly supervised setting.
We provide Python code for data management and some first experiments in weed species classification on our GitHub repository (https://github.com/grimmlab/MFWD).
In summary, the MFWD is a valuable resource for the scientific community, providing a comprehensive collection of high-quality images and curated data to advance research in weed detection and sustainable agriculture. It opens opportunities for the development of innovative computer vision algorithms to address some of the major challenges in weed management.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
What are SDG Topics?
An introduction to Sustainable Development Goals (SDGs) Topics and their role in highlighting sustainable development research.
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Genomics in freshwater and marine science
Publishing Model: Open Access
Deadline: Jul 23, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in