Maps, Networks and Mechanisms: A Framework for Integrating Knowledge, Modeling and Analysis for decoding Metastatic Breast Cancer
Published in Cancer, Protocols & Methods, and Mathematics
Our goal was to create a conceptual disease map capturing the signaling intricacies associated with TGFβ induced Epithelial Mesenchymal Transition (EMT) leading to Metastatic Breast Cancer (MBC). This work also explored the use of disease maps to comprehend the complexities of the MBC through various approaches such as discrete dynamic model simulation, network analysis, and transcriptome-based analysis. Figure 1 illustrates the overall approach followed for the development of the map and systematic analysis performed.
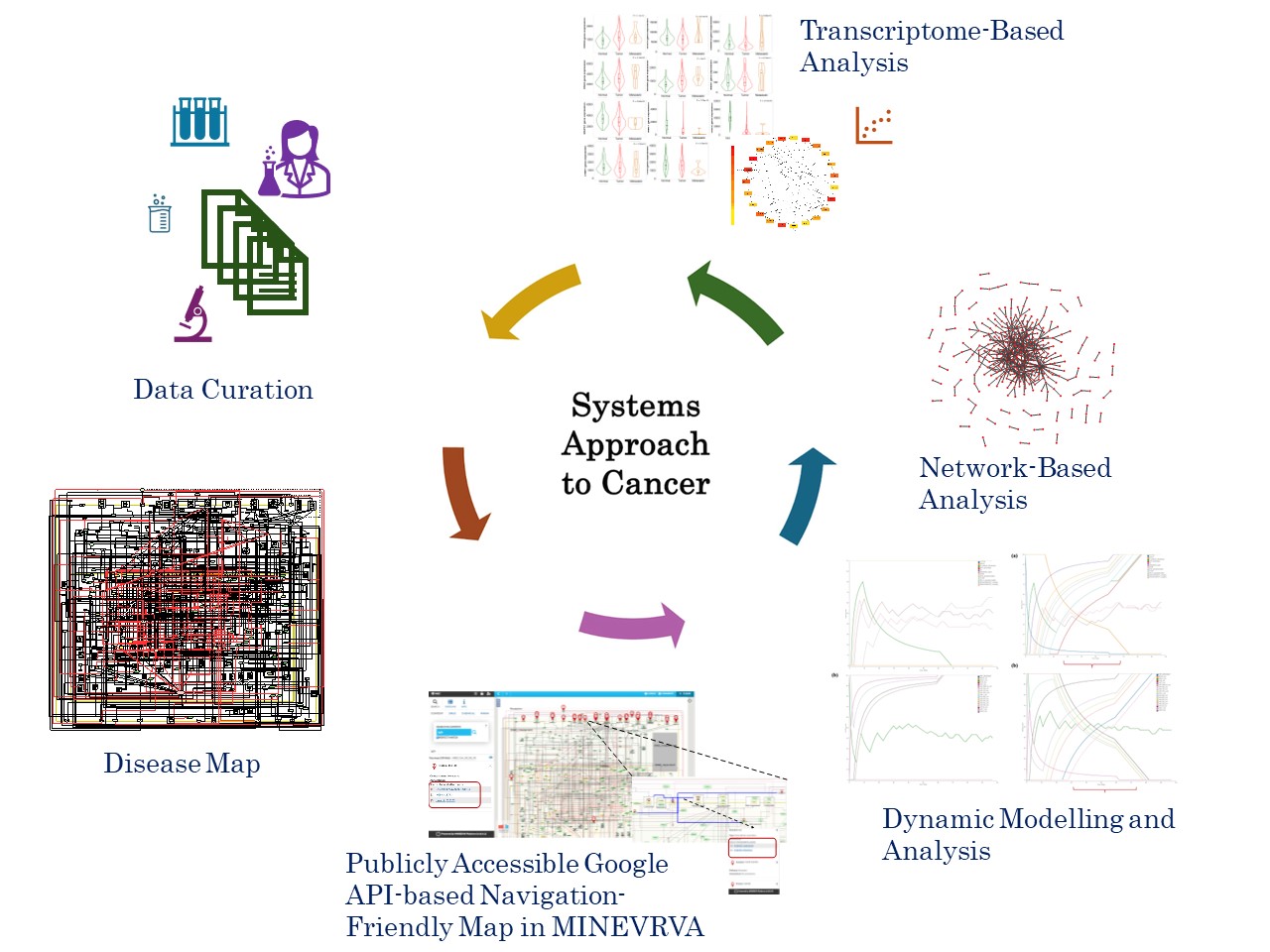
The heterogeneity of MBC, with its multitude of signaling pathways presents a significant challenge in cancer research and treatment. Cancer metastasis, a leading cause of cancer-related deaths worldwide involves cancer cells detaching from their primary sites, migrating, and invading distant tissues. EMT, a cellular transdifferentiation process where epithelial cells lose their adhesive nature, acquire migratory, and invasive properties (Brabletz et al., 2021). Numerous signaling pathways plays a crucial role in orchestrating this cellular transformation. The paradoxical role of transforming growth factor-beta (TGFβ) in cancer, both as a tumour suppressor and promoter, further complicates the process of TGFβ induced EMT in MBC. Understating the intricate web of signaling pathways in diseases like MBC is crucial for gaining deeper insights about the disease progression and to develop treatment strategies.
Disease maps serves as comprehensive, knowledge-based digital twin for complex diseases like MBC. These maps offer concise visualisation of how various regulators interact, highlighting the integrative role of signaling pathways and intricate cross-regulation. Analogous to a geographic map guiding exploration, disease maps empower researchers to navigate the complex molecular signaling landscape of a disease. By analysing network patterns within these maps, researchers can uncover the underlying network architecture and its functional significance. System-level models derived from disease maps can potentially reveal emergent properties, providing deeper insights into disease processes. In proliferative diseases like cancer, they can aid in tracking the disease progression at molecular level and to develop interventions strategically to target combinatorial pathways.
While embarking on the journey to build the disease map specific to TGFb induced EMT in breast cancer cell lines, we adopted a systemic effort to curate data from experimental literature on breast cancer cell lines. A preliminary version of the map developed was presented at the disease map community meeting DMCM -2021 (https://disease-maps.org/DMCM2021/ ). This forum provided valuable feedback which further helped in refinement of the map. The map underwent further development, resulting in the current version, strictly adhering to the systems biology standards and is publicly made available at MINERVA platform http://35.174.227.105:8080/minerva/?id=Metastatic_Breast_Cancer_1 for the research community. The effectiveness of such maps relies on the quality and reliability which is ensured by linking each regulator to their respective HGNC ID, uniport ID, Ensembl identifiers along with the PUBMED identifiers. We are grateful to Prof. Trivadi Sundaram Ganesan, a practicing clinical oncologist, whose insights and expertise from the clinical perspective has significantly improved this study.
With the feedback and suggestions from Prof. Anna Niarakis and in alignment with our objectives the MBC map developed was translated into a Boolean inference model using CaSQ and modelled using Cell Collective to validate its accuracy in replicating the curated experimental information. The model successfully replicated experimental results. Subsequent topological analysis of the assembled map revealed hub regulators implicated in the development of EMT in MBC and the transcriptome-based analysis validated the prognostic significance of these hub elements. Many of the hub regulators identified were EMT regulators, oncogenes, tumour suppressors and driver genes which are potential targets for therapeutic interventions.
Concurrently, we also conducted studies on the initial versions of the map to analyse simple topological metrics and gain insights into the architecture. These analyses revealed the presence of hierarchical network motifs, including coherent feed-forward loops, organized in a hypermotif architecture. Dynamic properties of Hypermotifs, explored by Adler & Medzitov (2022) (Adler & Medzhitov, 2022), and the classic work by Prof. Novikoff (1945) (Novikoff, 1945) on the interdependence and importance of hierarchical organisation both served as significant inspiration for our systems level studies. Our subsequent systems level mathematical modeling and analysis revealed their involvement in shaping the dynamics and functionality during disease progression (Bhavani & Palanisamy, 2022; Gottumukkala & Palanisamy, 2023; Sai Bhavani & Palanisamy, 2023).
Developing advanced medical digital twins, especially for complex diseases like MBC, presents significant challenges, but also holds great potential. One ongoing challenge is keeping the maps continuously updated as new data emerges. However, this can be addressed. Another challenge lies in moving beyond simply understanding these maps and to promoting their practical use in clinical decision-making. This may likely require interdisciplinary and collaborative efforts involving immunologists, clinicians, mathematical modelers, and software engineers to make substantial progress in effective utilisation of these maps. In future, by integrating patient-specific data, such as genetic profiles and tumour characteristics, into these digital twins, one can predict disease progression and treatment outcomes with greater accuracy.
Overall, comprehensive disease maps are powerful visual tools to explore molecular regulation or cross regulation of complex diseases like cancer. We hope that the map built in this study and the insights gained through the various analyses help the community to develop more effective strategies for understanding and treatment of complex diseases like cancer.
References
Adler, M., & Medzhitov, R. (2022). Emergence of dynamic properties in network hypermotifs. Proceedings of the National Academy of Sciences, 119(32). https://doi.org/10.1073/pnas.2204967119
Bhavani, G. S., & Palanisamy, A. (2022). SNAIL driven by a Feed Forward Loop Motif Promotes TGFβ Induced Epithelial to Mesenchymal Transition. Biomedical Physics & Engineering Express, 8(4). https://doi.org/10.1088/2057-1976/ac7896
Brabletz, S., Schuhwerk, H., Brabletz, T., & Stemmler, M. P. (2021). Dynamic EMT: a multi‐tool for tumor progression. The EMBO Journal, 40(18). https://doi.org/10.15252/embj.2021108647
Gottumukkala, S. B., & Palanisamy, A. (2023, 14-17 Dec. 2023). Cancer Networks: Insights Into The DiseaseMechanisms. 2023 IEEE 20th India Council International Conference (INDICON),
Novikoff, A. B. (1945). THE CONCEPT OF INTEGRATIVE LEVELS AND BIOLOGY. Science, 101(2618), 209-215. https://doi.org/10.1126/science.101.2618.209
Sai Bhavani, G., & Palanisamy, A. (2023). Network motifs and hypermotifs in TGFβ-induced epithelial to mesenchymal transition and metastasis [Original Research]. Frontiers in Systems Biology, 3. https://doi.org/10.3389/fsysb.2023.1099951
Follow the Topic
-
npj Systems Biology and Applications

An online Open Access journal dedicated to publishing the premier research that takes a systems-oriented approach and encourages studies that integrate, or aid the integration of, data, analyses and insight from molecules to organisms and broader systems.
Related Collections
With Collections, you can get published faster and increase your visibility.
Next-Generation Mammalian Cell Bioprocessing: Systems Biology, Synthetic Biology, and Beyond
Publishing Model: Open Access
Deadline: Mar 15, 2026
Systems mechanobiology
Publishing Model: Open Access
Deadline: Mar 28, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in