
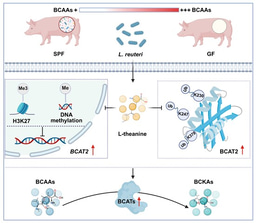
METZYME was a research expedition which aimed to connect trace metal nutrient distributions (MET) with microbial proteins and enzymes (ZYME) in the ocean – an endeavor which has enabled a powerful glimpse into the marine microbiology and biogeochemistry of the central Pacific Ocean. The goal of this expedition was to examine the role trace metals play in surface primary productivity and remineralization, ultimately improving our understanding of how the chemical environment influences biogeochemical cycling of carbon in the ocean. METZYME was a smashing success, providing insights into cyanobacterial nutritional stress and requirements (Saito et al. 2014, 2015; Hawco et al. 2020), deep sea chemoautotrophy (Santoro et al. 2017; Saito et al. 2020a), mercury and selenium biogeochemistry (Lamborg et al. 2014; Soerensen et al. 2014; Munson et al. 2015, 2018; DiMento and Mason 2017; Mason et al. 2018), and offshore dinoflagellate ecology and nutrient strategies (Cohen et al. 2021). METZYME also turned out to be a valuable exercise in being prepared and quickly adapting to unexpected events while at sea.

Sediment trap array being deployed to collect sinking particles and estimate carbon flux during the METZYME expedition. Photo credit: Mak Saito
As many oceanographers know, research cruises are a physically and mentally grueling experience. They involve months of logistical effort leading up to them to ensure appropriate sample collection, storage and transport across international lines, and require detailed knowledge of ocean instrumentation and engineering. The field was only just figuring out how to orchestrate multi-‘omic collection alongside trace metal clean seawater sampling when this expedition occurred. In the Saito lab, this process involves attaching ~80 pound McLane pumps to a winch line which gets lowered over the side of the ship, filtering hundreds of liters of seawater in situ to collect enough biomass for later ‘omic investigations, and setting up a makeshift trace metal clean “bubble” room onboard the ship (which is made almost entirely of metal) for the chemical analyses. Some of us (me) can barely carry an 8L niskin bottle full of seawater, let alone lift an 80 pound McLane pump! These operations are a tremendous group effort, and would not be possible without the hard work and collaboration of the ship crew and science team.

Makeshift trace metal clean room onboard R/V Kilo Moana for seawater collection from niskin bottles. Photo credit: Brian DiMento
The METZYME expedition was full of surprises, and actually almost failed early on. The entire CTD rosette equipped with niskin collection bottles and chemical sensors was lost to the bottom of the ocean at Station 3. A second CTD sensor package was purchased for the Trace Metal Clean rosette before the cruise, and this became the main sensor until replacement parts were sent over to Christmas Island from the University of Washington halfway through the expedition. In another stroke of bad luck, the counter block attached to the ships’s A-frame that tracks CTD depth broke, although thankfully the scientists brought an extra block onboard. One of the bigger surprises from this expedition was the relative abundance of dinoflagellates; they were prevalent across the transect in both surface and deep waters, yet functionally, their metabolic profiles were very different. Since dinoflagellates were not expected to be dominant contributors to the molecular pools, efforts to isolate live cells and bring them home were not made, which was unfortunate since little is known about offshore dinoflagellate physiology. Another shock was the discovery of a hydrothermal plume directly below the ship at the very last sampling location, which was only identified after analyzing seawater for trace metal content back in the lab. The science team had no idea at the time that the hydrothermal metal signal would be decoupled from the previous measurements of hydrothermal helium, and biomass was not collected at the heart of this metal plume, although microbes were almost certainly responding to this intense chemical feature. And due to the earlier delays, there was almost no time left to extensively sample at this station. Despite these setbacks and limitations, most of the intended oceanographic samples were successfully collected, and the results enhanced our understanding of trace metal distributions and microbial metabolism across the vast central Pacific Ocean.

Backup CTD parts arriving on Christmas Island via boat halfway through the expedition. Photo credit: Mak Saito
Oceanographers could spend years analyzing ocean samples collected from a single expedition crossing geochemical gradients and ecosystems (and we have!). METZYME is an especially valuable expedition in that a variety of chemical and biological parameters were simultaneously collected throughout the water column, producing millions of transcripts and thousands of proteins alongside nutrient concentrations, and enabling spatial mapping of marine metabolism across vertical and lateral ocean transects. There is still much to be discovered within this public dataset (Saito et al. 2020b). We hope it will be a valuable reference for other current and future marine biogeochemists, and may it also serve as a cautionary tale: bring two of everything on your next research expedition. In a fitting celebration on one of the last METZYME evenings, the ship’s chef prepared a backup chocolate cake – just in case.

Trace metal rosette being recovered after successful seawater sample collection. Photo credit: Brian DiMento
Cohen, N. R., M. R McIlvin, D. M. Moran, and others. 2021. Dinoflagellates alter their carbon and nutrient metabolic strategies across environmental gradients in the central Pacific Ocean. Nat Microbiol. https://doi.org/10.1038/s41564-020-00814-7
DiMento, B. P., and R. P. Mason. 2017. Factors controlling the photochemical degradation of methylmercury in coastal and oceanic waters. Mar. Chem. 196: 116–125. doi:10.1016/j.marchem.2017.08.006
Hawco, N. J., M. M. McIlvin, R. M. Bundy, and others. 2020. Minimal cobalt metabolism in the marine cyanobacterium Prochlorococcus. Proc. Natl. Acad. Sci. 117: 15740 LP-15747. doi:10.1073/pnas.2001393117
Lamborg, C. H., C. R. Hammerschmidt, K. L. Bowman, and others. 2014. A global ocean inventory of anthropogenic mercury based on water column measurements. Nature 512: 65–68. doi:10.1038/nature13563
Mason, R. P., A. L. Soerensen, B. P. DiMento, and P. H. Balcom. 2018. The Global Marine Selenium Cycle: Insights From Measurements and Modeling. Global Biogeochem. Cycles 32: 1720–1737. doi:10.1029/2018GB006029
Munson, K. M., C. H. Lamborg, R. M. Boiteau, and M. A. Saito. 2018. Dynamic mercury methylation and demethylation in oligotrophic marine water. Biogeosciences 15: 6451–6460. doi:10.5194/bg-15-6451-2018
Munson, K. M., C. H. Lamborg, G. J. Swarr, and M. A. Saito. 2015. Mercury species concentrations and fluxes in the Central Tropical Pacific Ocean. Global Biogeochem. Cycles 29: 656–676. doi:10.1002/2015GB005120
Saito, M. A., A. Dorsk, F. Anton, M. R. Mcilvin, M. S. Rapp, G. R. Ditullio, and D. M. Moran. 2015. Needles in the blue sea : Sub-species specificity in targeted protein biomarker analyses within the vast oceanic microbial metaproteome. Proteomics 0: 1–11. doi:10.1002/pmic.201400630
Saito, M. A., M. R. McIlvin, D. M. Moran, and others. 2020a. Abundant nitrite-oxidizing metalloenzymes in the mesopelagic zone of the tropical Pacific Ocean. Nat. Geosci. 13: 355–362. doi:10.1038/s41561-020-0565-6
Saito, M. A., M. R. McIlvin, D. M. Moran, T. J. Goepfert, G. R. DiTullio, A. F. Post, and C. H. Lamborg. 2014. Multiple nutrient stresses at intersecting Pacific Ocean biomes detected by protein biomarkers. Science (80-. ). 345: 1173–1177. doi:10.1126/science.1256450
Saito, M. A., J. K. Saunders, M. Chagnon, and others. 2020b. Development of an Ocean Protein Portal for Interactive Discovery and Education. J. Proteome Res. doi:10.1021/acs.jproteome.0c00382
Santoro, A. E., M. A. Saito, T. J. Goepfert, C. H. Lamborg, C. L. Dupont, and G. R. DiTullio. 2017. Thaumarchaeal ecotype distributions across the equatorial Pacific Ocean and their potential roles in nitrification and sinking flux attenuation. Limnol. Oceanogr. 62: 1984–2003. doi:10.1002/lno.10547
Soerensen, A. L., R. P. Mason, P. H. Balcom, D. J. Jacob, Y. Zhang, J. Kuss, and E. M. Sunderland. 2014. Elemental mercury concentrations and fluxes in the tropical atmosphere and Ocean. Environ. Sci. Technol. 48: 11312–11319. doi:10.1021/es503109p
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in