Microbial thieves from the barnyard
Published in Microbiology
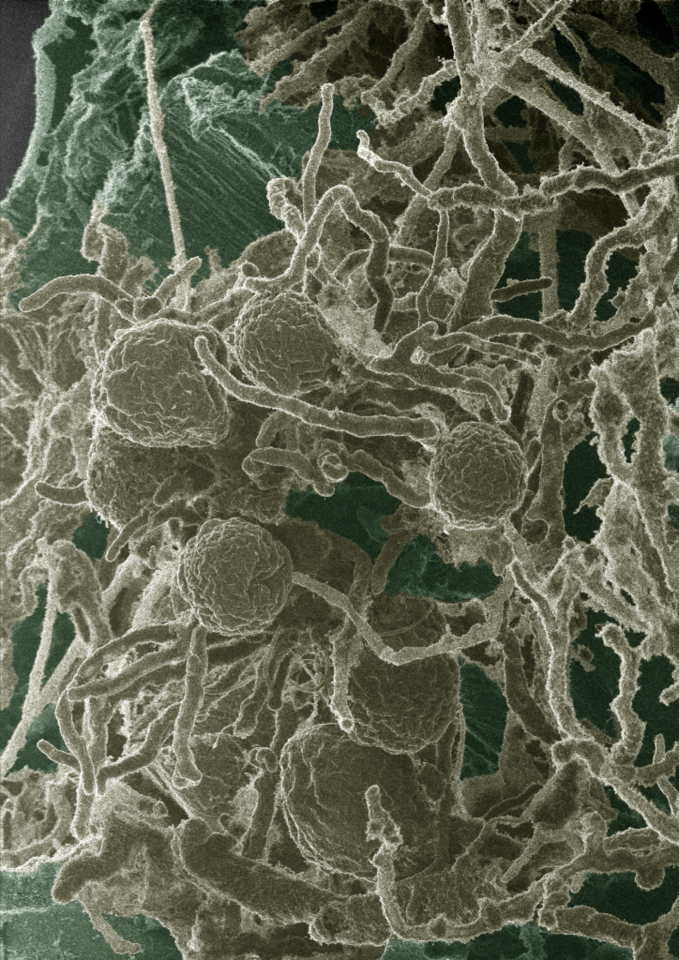
When trying to solve difficult problems in engineering, one solution is to turn to nature for inspiration. In the rumen or hindgut of large herbivores, a diverse population of microbes enables rapid conversion of plant material. Among them, anaerobic fungi are remarkably robust at breaking down plant material. Our lab went straight to the source to isolate and characterize fungi from fecal samples at the Santa Barbara Zoo. The zoo is more than happy to give us “inspiration”, often by the dumpster-load.

One of the ways in which the fungi achieve efficiency in degrading plant material is through the production of large, multi-protein complexes called cellulosomes. For over thirty years, the research community has reported the existence of cellulosomes in anaerobic fungi, however their mechanism for assembly, and specifically the scaffolding protein onto which the cellulases assemble has remained elusive. A similar system has been characterized in anaerobic bacteria, but the parts have no similarity.
Our lab performed a few different biochemical assays to try to pin down what the cellulases were sticking to, without many definitive results. The real breakthrough came when we employed proteomics as part of a multi-omics study to acquire the genomes, transcriptomes, and proteomes of three novel isolates of anaerobic fungi in collaboration with the Joint Genome Institute (JGI) and the Environmental Molecular Sciences Laboratory (EMSL). The proteome (acquired at EMSL) mapped to the transcriptome (acquired at JGI) revealed several large (~700 kDa), completely uncharacterized proteins in both the secretome and cellulose-bound fraction of the anaerobic fungi. Our biochemical assays at UCSB then verified these proteins as the scaffolding proteins, answering the 30-year question as to how the cellulosomes of anaerobic fungi assemble.

In our work, we also report the highest quality genomes assembled for anaerobic fungi to date. The genomes are extremely AT-rich, with the highest AT content at 84% in Anaeromyces robustus. The AT-richness previously led to problems in sequencing and assembly, with the best prior attempt at sequencing and assembly resulting in greater than 17,000 contigs. Leveraging the long reads generated by PacBio sequencing, we were able to assemble our highest quality genome into just 232 contigs. The high-quality genomes allowed for a more complete “parts list” for the fungal cellulosomes, including 95 of the newly identified scaffoldins and close to 1600 dockerin domain proteins (proteins with domains that interact with the scaffoldin).
Looking more closely at the dockerin proteins, many of them appeared to be bacterial in origin. Through a phylogenetic analysis, 10 of the catalytic domains on dockerin domain proteins were found to have undergone horizontal gene transfer between the anaerobic fungi and their prokaryotic neighbors. Even more interesting was that only the catalytic domain appeared to have been transferred, and any accessory domains also found were somehow added or removed after transfer.
The results of our work present for the first time a parts list of proteins in anaerobic fungal cellulosomes. The interacting domains bear no similarity to the system employed by anaerobic bacteria, suggesting that the importance of cellulosomes in breaking down plant material led to the co-evolution of separate systems in bacteria and fungi. Several catalytic domains appear to have arisen from gene transfer, resulting in an evolutionarily chimeric structure – an independently evolved structure with useful enzymes co-opted from bacterial neighbors.
Read more about our story at Nature Microbiology: http://go.nature.com/2sMEyl4





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in