
By Benjamin Winer, Gabriel Lipkowitz and Alexander Ploss
Chronic hepatitis B virus (HBV) infections are a global health burden afflicting approximately 250 million individuals around the world. If left untreated, chronic HBV can lead to cirrhosis, fibrosis, and hepatocellular carcinoma (HCC), a common type of liver cancer. Every year, approximately one million individuals succumb to complications of chronic HBV infection. Treatments that suppress viral production exist, but they rarely cure the patient.
A major obstacle to the identification and development of curative therapeutics for HBV is the highly restricted host and tissue tropism of the virus. HBV only productively infects human and chimpanzee hepatocytes (liver cells). Furthermore, human hepatocytes are notoriously difficult to culture as they de-differentiate quickly after plating, losing their susceptibility to HBV infection. Several groups have been working to extend the life span of primary human hepatocyte (PHH) culture systems, and great progress has been made over the last few decades. The development of co-cultures of PHHs with non-parenchymal mouse cells, and more recently of micro-patterned co-cultures (MPCCs), has prolonged the phenotype and functional characteristics of PHHs from a few days to up to a few weeks. However, HBV infections was still limited to just about 2-3 weeks, arguably modeling primarily the acute but not chronic phase of the infection Moreover, susceptibility to HBV was highly donor dependent and in order to achieve productive HBV infection in these systems, innate immunity must be blunted. Finally, HBV infection had not yet been reported in formats amenable to high-throughput drug screening.
Our novel, self-assembling co-culture primary human hepatocyte (SACC-PHH) platform overcomes these drawbacks. We demonstrate that SACC-PHH support persistent infection with cell culture and even patient derived HBV strains for 5-6 weeks without the need of antagonizing cell-intrinsic innate immunity. By resolving these technical issues, our system opens up several exciting avenues of studying HBV and conceivably other hepatotropic pathogens that were not tenable or very difficult in current PHH culturing platforms.
One such avenue of investigation is whether HBV, thought to be a stealth virus, activates the host’s innate immune responses in infected hepatocytes. If so, what specific genes are activated? If not, how does HBV evade the host defenses? Can host DNA sensing molecules detect the HBV genome? If not, how does the virus circumvent detection? Answers to these questions would give us a better understanding of the virus’s evasion mechanisms, thereby allowing us to possible develop innate immunotherapy treatments.
Another pressing question in the field of HBV research is when and how HBV genes integrate into the host genome. Finding the answers to these questions is critical, since viral integration is thought to promote the development of liver cancer. About 80% of all liver cancer cases globally are caused by chronic HBV infections. Now, using our robust SACC-PHH system, scientists can identify when integration events occur during HBV infection in a physiologically relevant cell type and elucidate the integration mechanisms used by the virus.
An additional, practical advantage of the SACC-PHH platform is its scalability. Unlike previous cell culture systems, our platform can be used in 96 well plates. As a result, it can support large screens for novel small molecule inhibitors of HBV infection in the native permissive host cell. Using our platform, researchers could test the anti-viral efficacy of candidate drugs, as well as quantify their metabolism and toxicity.
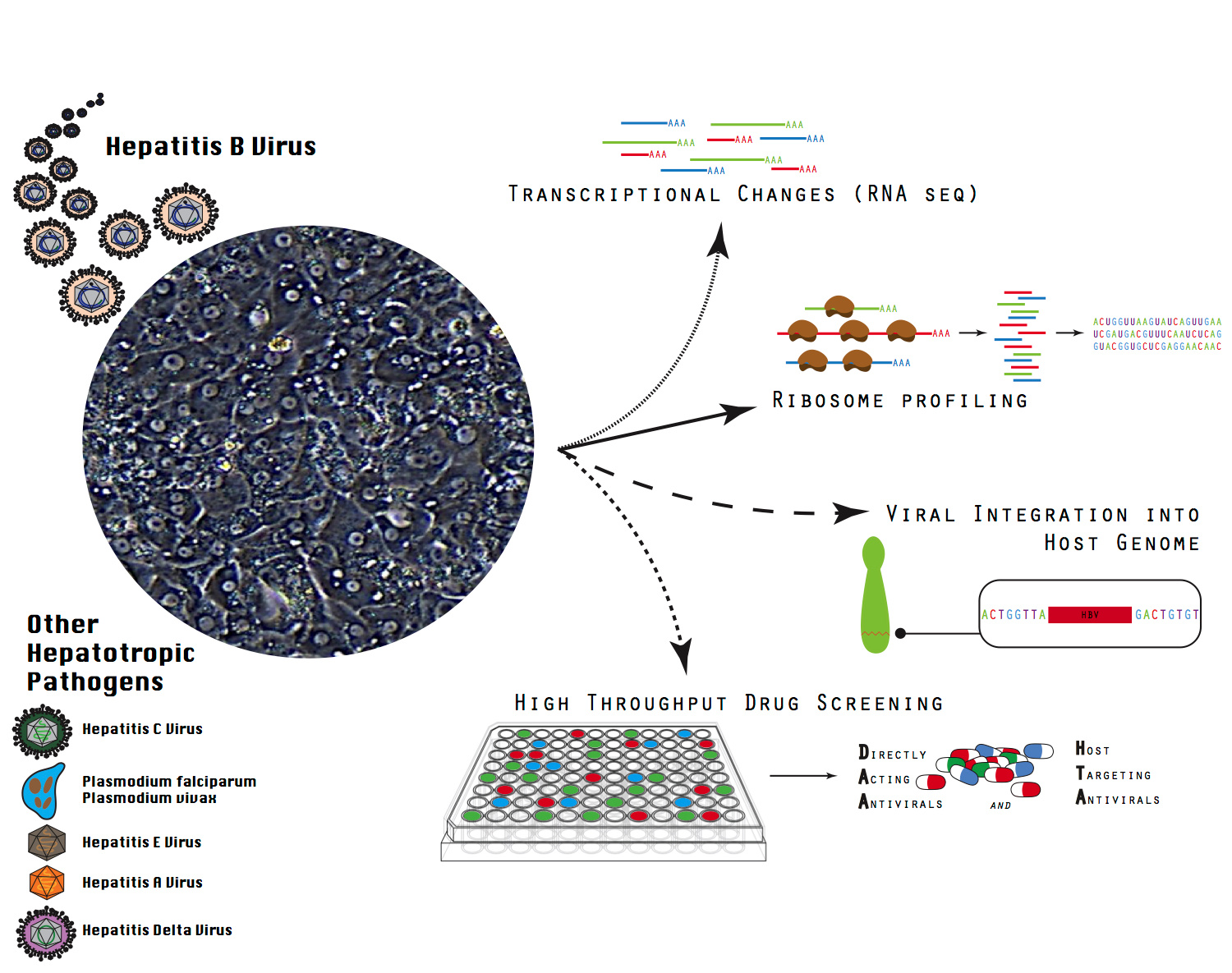
Figure 1: Schematic of potential new avenues of research utilizing the SACC-PHH platform to study hepatitis B virus.
More avenues of research are shown in the accompanying figure in this post.
This project is an excellent example of how an academic-industry collaboration, in this case between Princeton University and Hurel Corporation, can result in the development of novel scientific tools that can be broadly used in many different fields.
The paper can be found here: https://www.nature.com/article...





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in