New proteins on the test track
Published in Ecology & Evolution

The link to the paper in Nature Ecology and Evolution, published on March 19 2018, is here.
This paper delves into novel aspects of the formation of new genes-not about the kind of genes that arise by gene duplication, and thus are similar to other genes- but about genes which encode proteins with completely new sequences. Genomes are full of them, although we are only beginning to understand their evolutionary significance.
Genes that encode completely new proteins are called de novo genes. Comparisons of genomic sequences from closely related species support the idea that they are formed from previously non-coding sequences in the genome. A number of genes involved in spermatogenesis or brain functions may have originated de novo.
Initially, de novo genes arise by accident; in this sense, they are no different from gene duplicates. This may happen, for example, because RNA polymerase recognizes weak promoters that appear by chance in the genome, and initiates transcription of a previously silent segment of DNA.
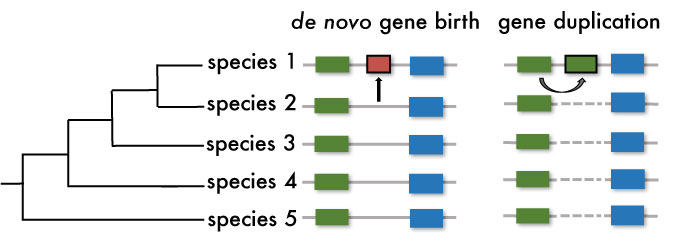
De novo gene birth versus gene duplication. In de novo gene birth a new gene (red box) emerges from a non-genic region in the lineage leading to species 1. In constrast, in gene duplication a new copy of an existing gene is generated (green boxes).
The majority of recently originated transcripts are unlikely to be of any use. Consequently, they will tend to disappear over time. However, as happens with gene duplicates, a few of them may turn out to be useful and be retained by natural selection. How likely it is that a random protein becomes functional? Nobody knows this for certain, but as Keefe and Szostak showed in 2001, some proteins will display certain biochemical activities just by chance if the number of starting proteins is large enough.
Random proteins are believed to have existed in the ‘primordial soup’ at the beginning of life, but do they still exist today? In our article, we identify many small proteins that appear to be translated by accident. They evolve under no constraints AKA neutrally, providing a constant supply of new peptides to be ‘tested’ for new functions.

José Luis Villanueva-Cañas (front) and Jorge Ruiz-Orera (back) analyzing data.
This work has unfolded over several years. When Jorge Ruiz-Orera, the first author of the study, arrived at the lab, we knew that the genome was pervasively transcribed, and that many of the transcripts were poorly conserved across species. Were they putative precursors of new protein-coding genes? We first had to determine if they could encode proteins, even if only short ones. Jorge used ribosome profiling data, a RNA sequencing technique that only targets actively translated regions, to search for evidence of translation. We found that thousands of mouse transcripts outside annotated protein-coding genes appeared to translate small proteins.
However, we still needed to know if these peptides were functional or not. On the one hand, these peptides could represent incorrectly-annotated protein-coding genes. On the other hand, they could be putative precursors of de novo genes. Thankfully, the Tautz lab had just published some mouse polymorphism data that we could use to identify signatures of purifying selection in non-conserved proteins. The problem was that the proteins were too small to do a one-by-one analysis. Then we realized that by using the codon composition of the sequences, it was possible to separate out a fraction of proteins that evolved under no selection from the rest of proteins. The first group had the expected features of de novo gene precursors- eureka!
The study has shown that there are many peptides on the "test track". Much work is left to do before we fully understand the impact of de novo genes in recent evolution, but the road ahead seems to be clearing up.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in