Polyploidy is a frequent phenomenon in nature, and its implications for genetics and agriculture are fascinating.
Published in Ecology & Evolution, Genetics & Genomics, and Agricultural & Food Science

We extensively use cytogenetics and genomic approaches to address fundamental questions in genetics and agriculture. Polyploidy is a frequent phenomenon in nature, and its implications for genetics and agriculture are fascinating; however, the underlying mechanics are complex and not fully understood.
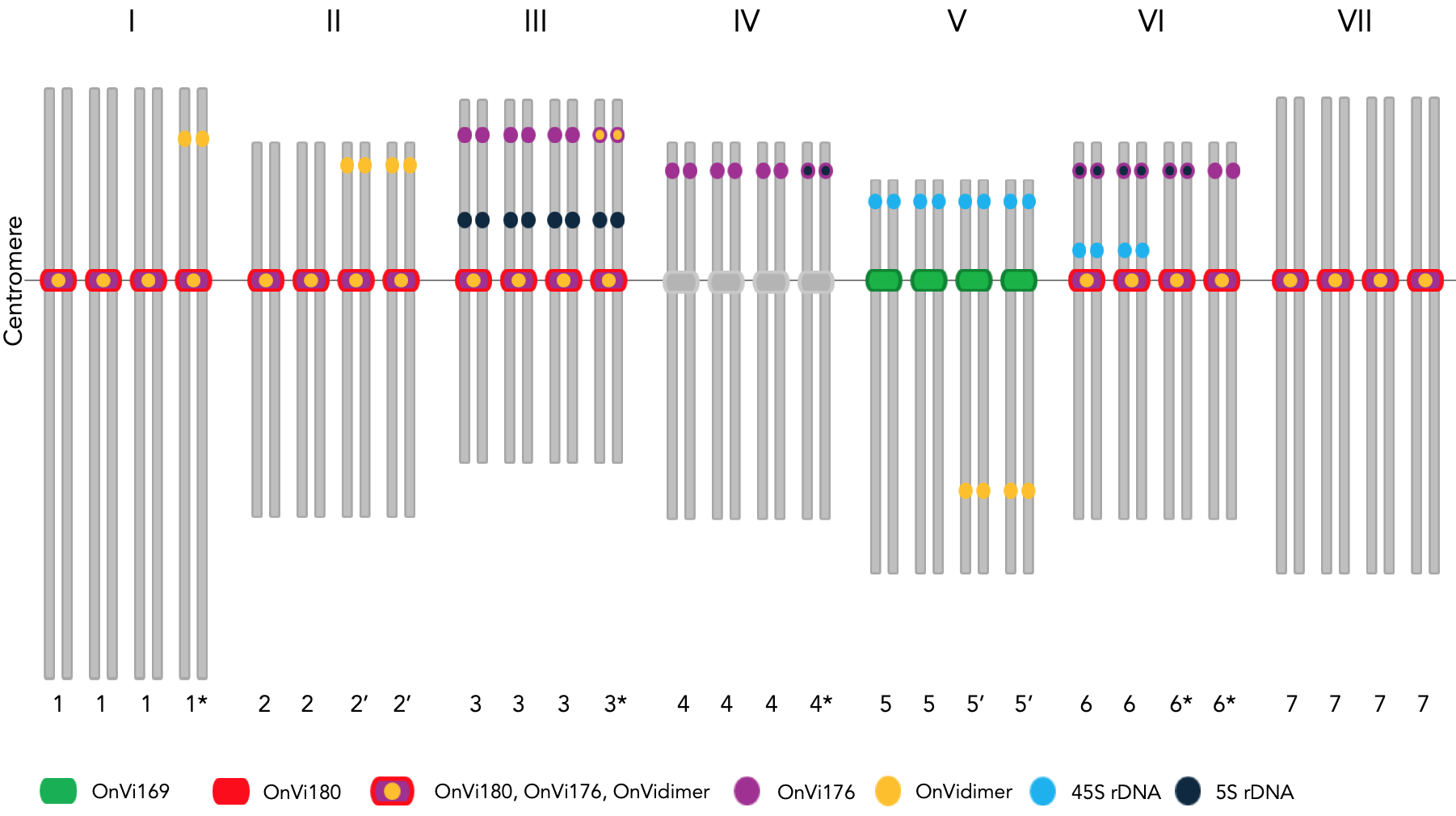
Polyploidy is a common feature in eukaryotes, and one of the paramount consequences is better environmental adaptation. Many plants, such as wheat and potato, are polyploids with enhanced adaptability to diverse environments. Sainfoin, Onobrychis viciifolia Scop., is a tetraploid (2n = 4x = 28, 1225 Mbp) forage legume plant belonging to the Fabaceae family with great potential in sustainable agriculture (Figure 1).

Traditionally, heterochromatin forms compact and heavily stained domains in eukaryotic genomes. Tandem repetitive DNA sequences are inherently either difficult to sequence or challenging to assemble. Even whole-genome sequencing approaches with next-generation sequencing technologies have limitations in resolving these genomic regions. We utilized a combination of cytogenetic, genomic and bioinformatic approaches to establish a comprehensive karyotype of sainfoin, define its heterochromatic chromosome regions and interpret its polyploid origin (Figure 2). Some questions that we raised in our study are whether sainfoin, an agriculturally successful example, can be a model for domestication and crop improvement, what we see in the sainfoin karyotype could be a hint of evolutionary consequences of polyploidy, and how the genome might be reorganized after genome duplication. Finally, we discuss our novel and complementary findings in this underutilized polyploid crop with unique and complex chromosomal features. Curiously, our conclusions may need to be tested and challenged in time with more advanced techniques.
We are a group of highly enthusiastic, motivated scientists who believe in the power of genetics and agriculture with a focus on chromosomes. We believe that our research experience in the laboratory is unique to our personal, scientific, and professional development. The support among the lab members at all levels, one-for-all, makes us achieve challenges in plant chromosome research to become scientists in the future. You can follow us on Instagram.
Our research is published in the prestigious journal Chromosoma under the Editorial Board members Ingo Schubert and Yamini Dalal. If you want to read our work, follow the link above. Please feel free to contact me at altek2@gmail.com if you have any questions or comments about the work.
Follow the Topic
-
Chromosoma

This journal publishes research and review articles on the functional organization of the eukaryotic cell nucleus, with a particular emphasis on the structure and dynamics of chromatin and chromosomes; the expression and replication of genomes; genome organization and evolution, and more.
Related Collections
With Collections, you can get published faster and increase your visibility.
Mechanobiology in the Nucleus
Publishing Model: Hybrid
Deadline: Ongoing
40 years of CENP-A
Publishing Model: Hybrid
Deadline: Ongoing


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in