
PhD student Tomohito Yamada & Professor Hideaki Tsuge.
Some pathogenic species of Clostridium employ the classic enzymatic “AB” binary protein toxins for poisoning cells. Clostridium perfringens, C. difficile, C. spiroforme, and C. botulinum all use similar binary toxins (iota toxin (Ia and Ib), CDT (CDTa and CDTb), CST (CSTa and CSTb), and C2 toxin (C2I and C2II), respectively). They consist of the enzymatic ‘A’ component, an actin-specific ADP-ribosyltransferase and the ‘B’ component that binds to the host cell and forms a membrane-spanning pore that functions as the translocation channel for each enzymatic component. The ’B’ component translocates the ‘A’ component into the host cell via the membrane in the acidic endosome.
In contrast, the Bacillus anthracis species uses a different binary toxin, which consists of two enzymatic proteins: the lethal (LF) and edema (EF) factors, and a protein translocation channel, PA. The PA heptameric pore structure was revealed to have extremely narrow φ-clamp passageway and a long membrane-spanning channel. However, a high-resolution structure of the enzyme-bound translocation channel has not been revealed among all the binary toxins.
We have studied the function and structure of the iota toxins, Ia and Ib. To understand the protein translocation mechanism,it is important to elucidate the structure of the Ib-pore as well as the Ia-bound Ib-pore. However, it was difficult to study the Ib-pore structure because it is a super-macromolecule assembly that is membrane-embedded and might be a mixed state of prepore and pore, therefore difficult to purify homogeneously. Recent cryo-EM studies have paved the way to elucidating the Ib-pore structure, as well as the Ia-bound Ib-pore structure.
Ib oligomerizes poorly at 37°C under acidic conditions following cleavage of the 20 kDa N-terminal propeptide. However, we found efficient oligomerization method through testing many conditions. This improvement in sample preparation allowed imaging of the Ib-pore and Ia-bound Ib-pore in two different states (the short stem and long stem) by cryo-EM at atomic resolution (2.8~2.9 Å resolution).
The Ia-bound Ib-pore structure shows that the N-terminal domain of a single Ia molecule binds to the Ib-pore. Notably, the complex structures show that the Ia N-terminus unfolds and leads to the narrowest φ-clamp by binding to the Ib-pore. For the first time, these structures provide valuable insight into how Ia would be translocated via a φ-clamp with an inner diameter of 6 Å. Compared to the LF-bound PA-pore model, the complex structure of the Ia-bound Ib-pore showed a unique binding mode and unfolding and translocation mechanisms even though they use similar translocation pores.
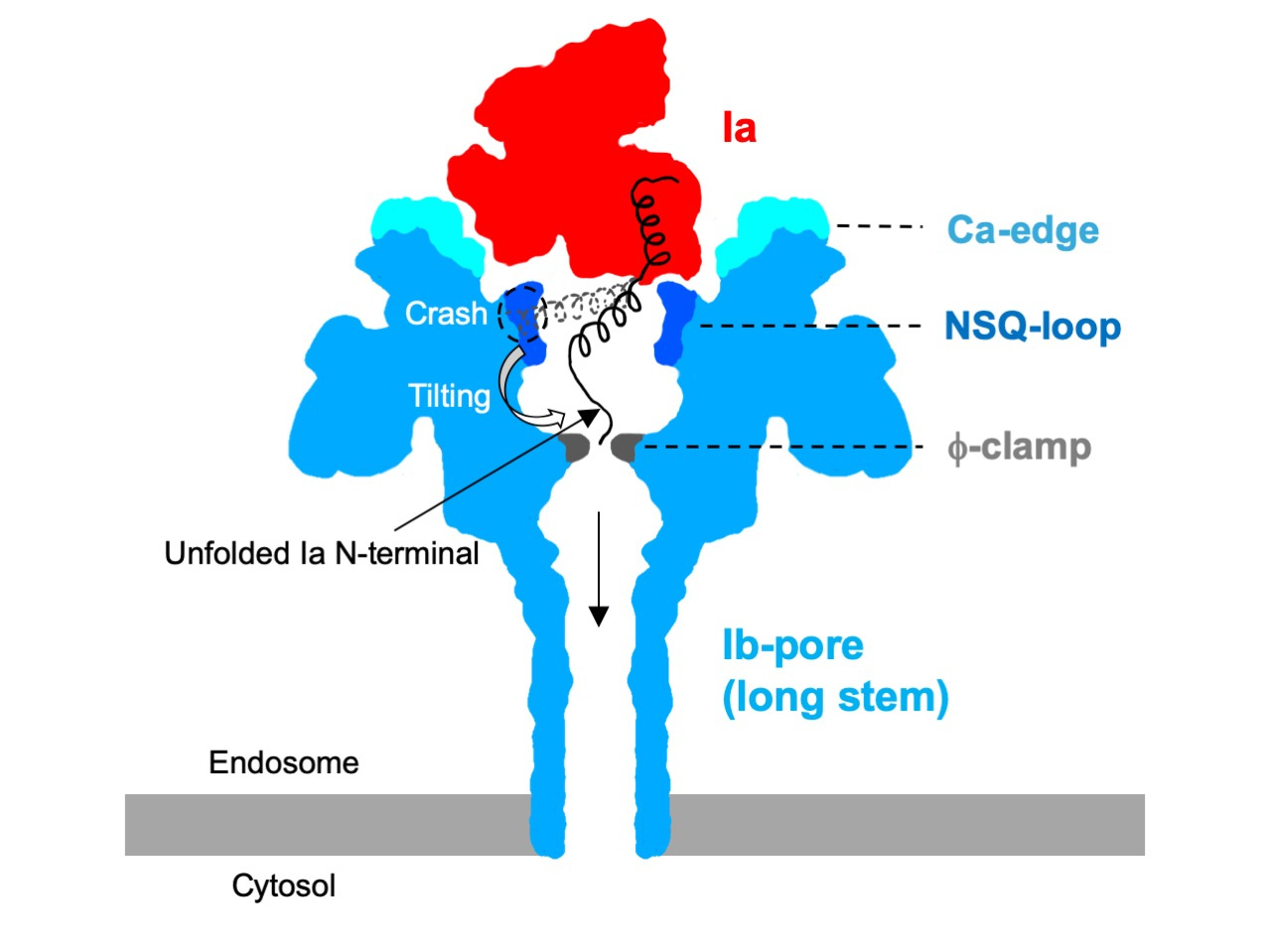
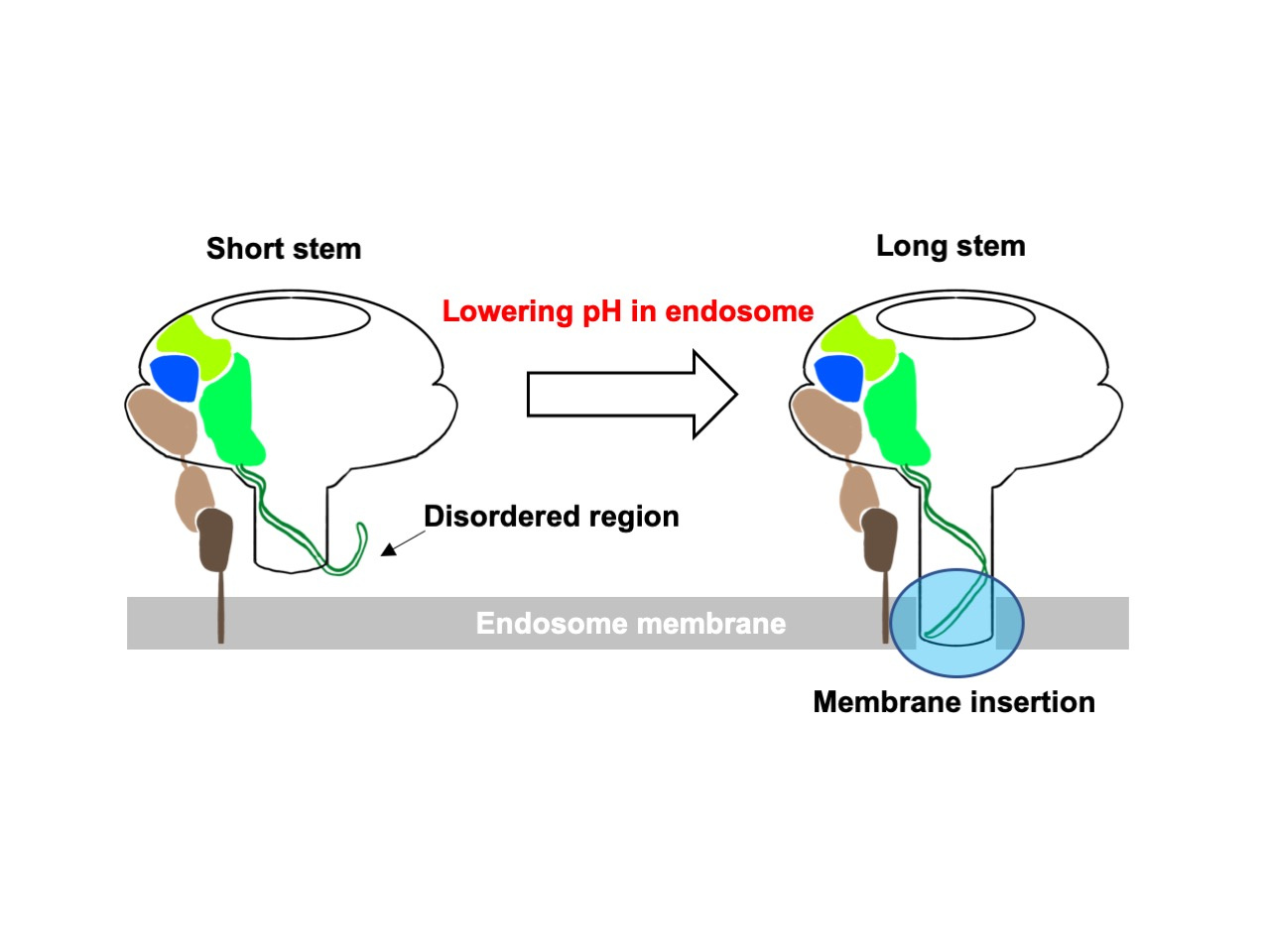
It is also noteworthy that two structures of the Ia-bound Ib-pores with short and long stems were obtained by cryo-EM 3D classification. The transmembrane region is disordered in the short stem structure. We hypothesize that Ib-pores with short stems are soluble and metastable intermediate states between the prepores and pores. The disordered region likely forms the intact beta-barrel (the long stem) from the short stem pore for membrane insertion, which may be general in the beta-pore-forming toxin. Recently, two groups reported that CDTb-pores form double heptamers, which is likely to be a non-physiological form, but they also include short stem pores with a disordered stem region1,2.
Check the full paper here.
1. Anderson, D.M., Sheedlo, M.J., Jensen, J.L. & Lacy, D.B. Structural insights into the transition of Clostridioides difficile binary toxin from prepore to pore. Nat Microbiol(2019).
2. Xu, X. et al. Structure of the cell-binding component of the Clostridium difficile binary toxin reveals a di-heptamer macromolecular assembly. Proc Natl Acad Sci U S A117, 1049-1058 (2020).
Follow the Topic
-
Nature Structural & Molecular Biology

An integrated forum for structural and molecular studies, the journal places a strong emphasis on functional and mechanistic understanding of how molecular components in a biological process work together.
Related Collections
With Collections, you can get published faster and increase your visibility.
DNA repair and human disease
Publishing Model: Hybrid
Deadline: Apr 30, 2026
Artificial Intelligence Methodology in Structural Biology
Publishing Model: Hybrid
Deadline: May 31, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in