ProHap: A New Tool for Capturing Genetic Diversity in Proteomics
Published in Protocols & Methods, Genetics & Genomics, and Biomedical Research
The field of genomics has made remarkable progress with the advent of human population-wide genetic reference panels. These have enabled mapping haplotypes — combinations of alleles at different places on the chromosome that are inherited together — across various populations. However, while genomics provides insights into the genetic blueprint, it alone cannot capture the full complexity of biological systems. Proteomics, the large-scale study of proteins, offers a deeper understanding of how genes are expressed and function in the body. Despite its potential, proteomics has so far faced challenges in accounting for genetic diversity.
The Challenge in Proteomics
Proteomic studies focus on the sequence, structure, function, and interactions of proteins - the workhorses of the cell. Proteins are directly involved in most biological processes and are often the targets of drugs. Therefore, understanding the proteome is crucial for uncovering the mechanisms of diseases, identifying biomarkers, and developing new therapies. Mass spectrometry-based analysis is the reference platform for proteomics, allowing scientists to identify and quantify peptides at scale in complex biological samples.
Mass spectrometry-based protein analysis often relies on reference sequences that do not represent the genomic diversity in the world’s populations. This oversight can lead to biases, particularly against populations that are underrepresented in reference databases.
Introducing ProHap
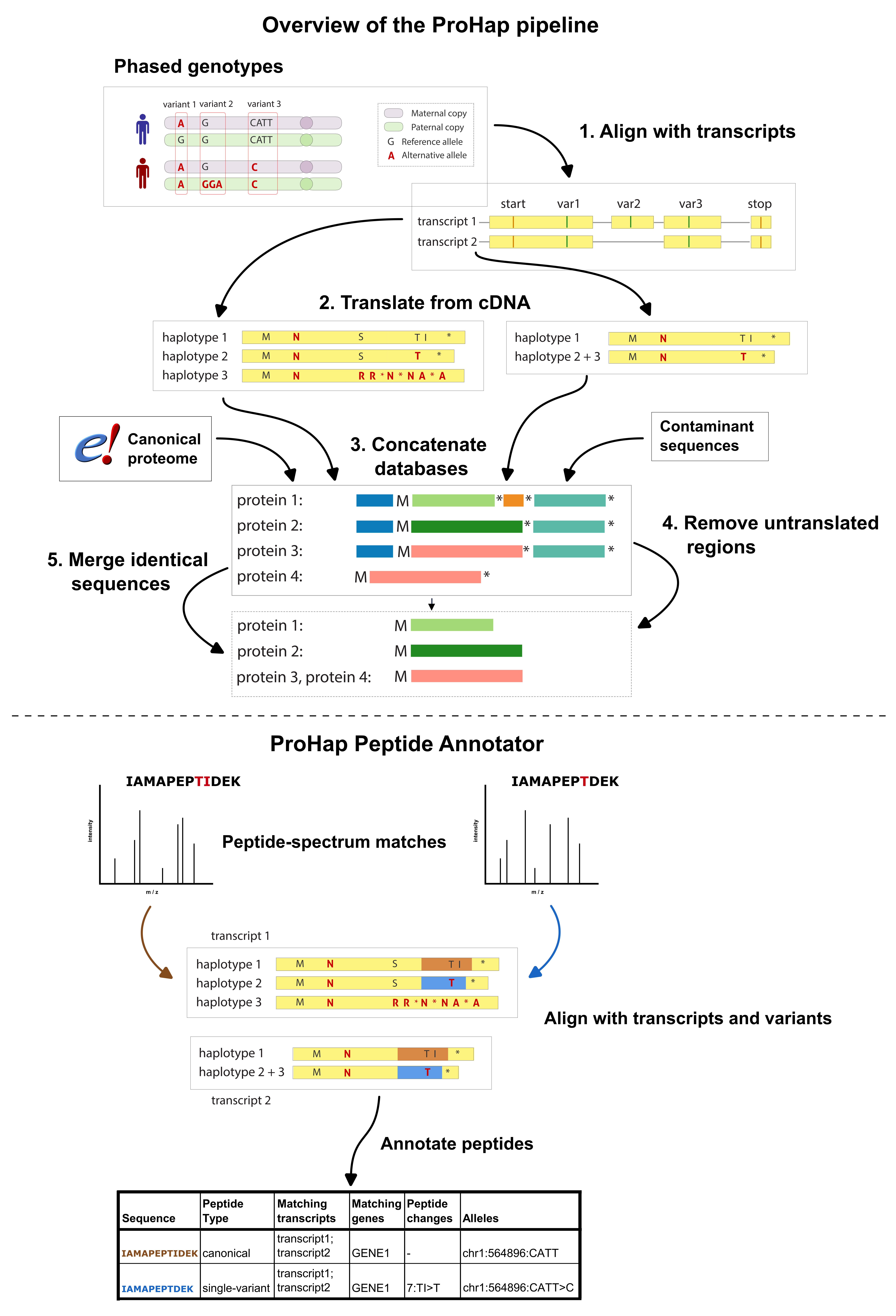
ProHap is a bioinformatic tool designed to address these challenges by creating protein sequence databases from large reference panels of human haplotypes. It uses phased genotypes to construct protein sequences possibly encoded by these haplotypes. Conversely, once a set of peptides is obtained from a proteomic search engine, the ProHap Peptide Annotator maps the peptides to their corresponding genes, haplotypes, and transcripts. Both tools are open-source, with code and documentation available in GitHub: https://github.com/ProGenNo/ProHap, https://github.com/ProGenNo/ProHap_PeptideAnnotator.
Ready-to-use protein sequence databases
To showcase ProHap's utility and provide resources to the scientific community, we used the tool to construct protein sequence databases using phased genotypes from three different sources:
- The 1000 Genomes Project (https://doi.org/10.5281/zenodo.10149277),
- Haplotype Reference Consortium Release 1.1 (https://doi.org/10.5281/zenodo.12671301),
- The first release of the Human Pangenome Reference Consortium (https://doi.org/10.5281/zenodo.12686818).
These databases are publicly available, and can be used with popular search engines, such as MaxQuant, MSFragger, X!Tandem, Tide, or DIA-NN.
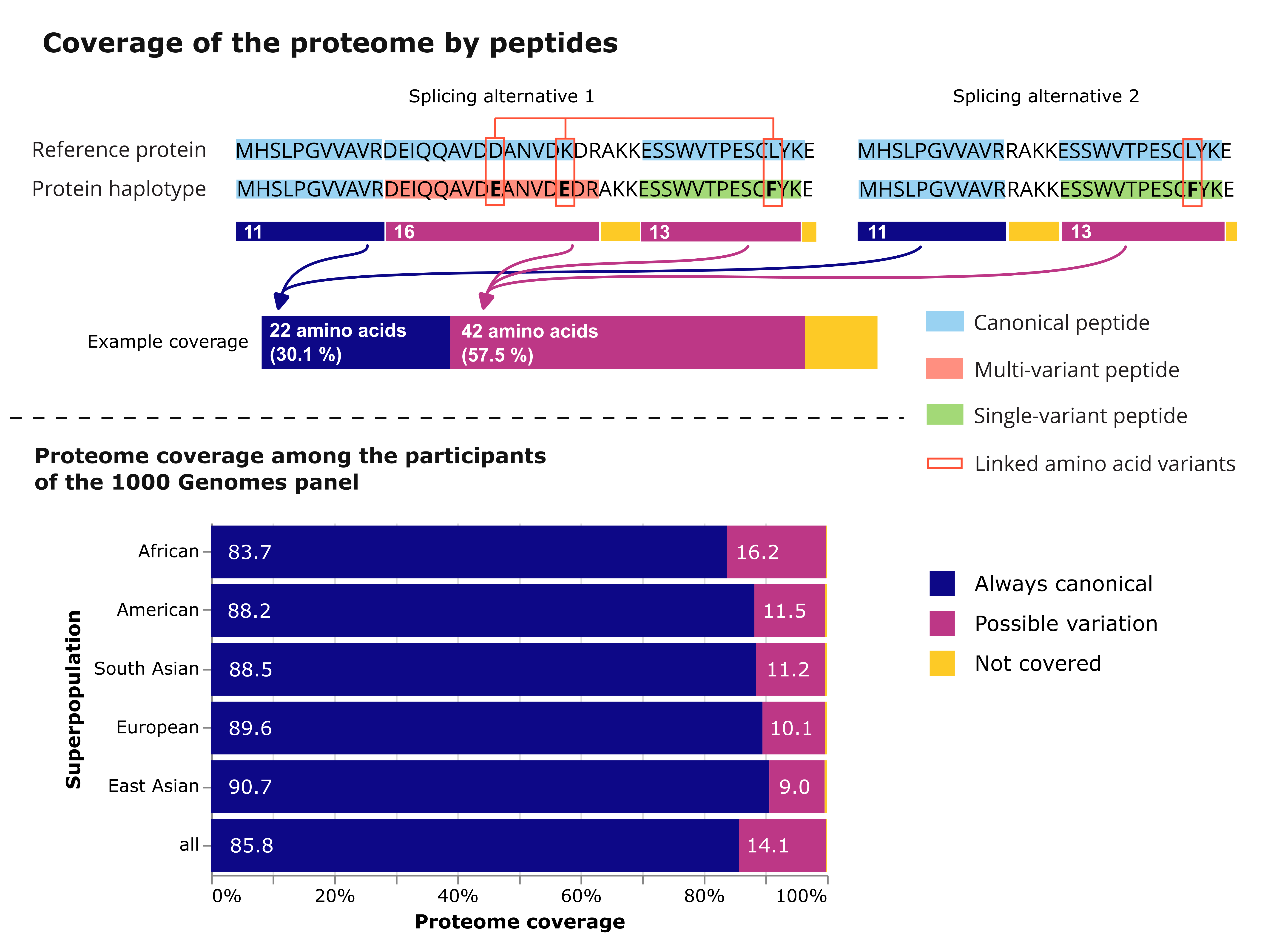
Our analysis of the databases created using the 1000 Genomes Project dataset revealed that a higher share of the proteome is affected by common haplotypes in participants from the African superpopulation. The proteomes of all the five included populations - African, American, European, South Asian and East Asian - show that over 9 % of all amino acid residues map to peptides that may carry a product of an alternative allele. This further underlines the need for integrating genetic diversity into proteomic workflows. Publicly available datasets, such as the 1000 Genomes, only partially capture human genetic diversity, while access to genomic data of indigenous populations is often restricted, recognizing community rights and interests. As a standalone pipeline, ProHap can be executed on secure servers using own panels of phased genotypes, maintaining the ownership of and access to the data.
Implications for proteomics
By providing a more inclusive and accurate representation of the proteome, ProHap enables researchers to better understand the genetic basis of diseases and potentially develop more effective, personalized treatments. This tool also highlights the importance of including diverse populations in proteomic research.
However, accounting for common variation also uncovers challenges that are invisibilized using reference sequences alone. For instance, it is a well-known issue that inflation in the size of sequence databases causes the search engine to return more false positives, in turn yielding a smaller number of peptide identifications at the same false discovery rate (FDR) threshold. The low number of new peptides introduced by common variants does not dramatically increase the search space, but by nature, the peptides encoded by different haplotypes present a high similarity - and the problem becomes increasingly complex due to the possibility to confuse a peptide encoded by an allele and a modified peptide encoded by another.
In proteomics, error rate estimation is based on a null distribution of scores modeled with random (decoy) matches. These are therefore not suited to track mismatches between peptides that are only partially incorrect (i.e., better than random, but still wrong). When working with reference sequences only, peptides originating from alternative haplotypes are not matched or incorrectly matched to a resembling sequence without being tracked by error rates, and the problem is ignored. How do we account for haplotypic variation in protein inference? How do we improve error rates to distinguish resembling peptides? These questions need to be resolved before proteomics can routinely tackle human diversity.
Moreover, most studies involving proteomics use summarization techniques to aggregate the abundance of hundreds of thousands of peptides into protein abundance estimates. If we introduce a number of similar protein sequences encoded by different haplotypes, we may expect different “versions” of the same protein in the same sample, or across different individuals of the same cohort. Currently, standard protein quantification pipelines do not account for sequence variation. Yet, the abundance of proteins is going to vary depending on the genome of the individual. Given the abundances of all the identified peptides, what is the difference between the abundances of a protein encoded by the maternal vs. the paternal haplotype? The ability to distinguish protein haplotypes provides a unique opportunity to increase the depth of our knowledge at the interface of genomics and proteomics.
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
A nice tool! Congratulations!
Thanks!