Prototyping an Ontological Framework for Cellular Senescence Mechanisms: A Homeostasis Imbalance Perspective
Published in Protocols & Methods and Cell & Molecular Biology
Introduction
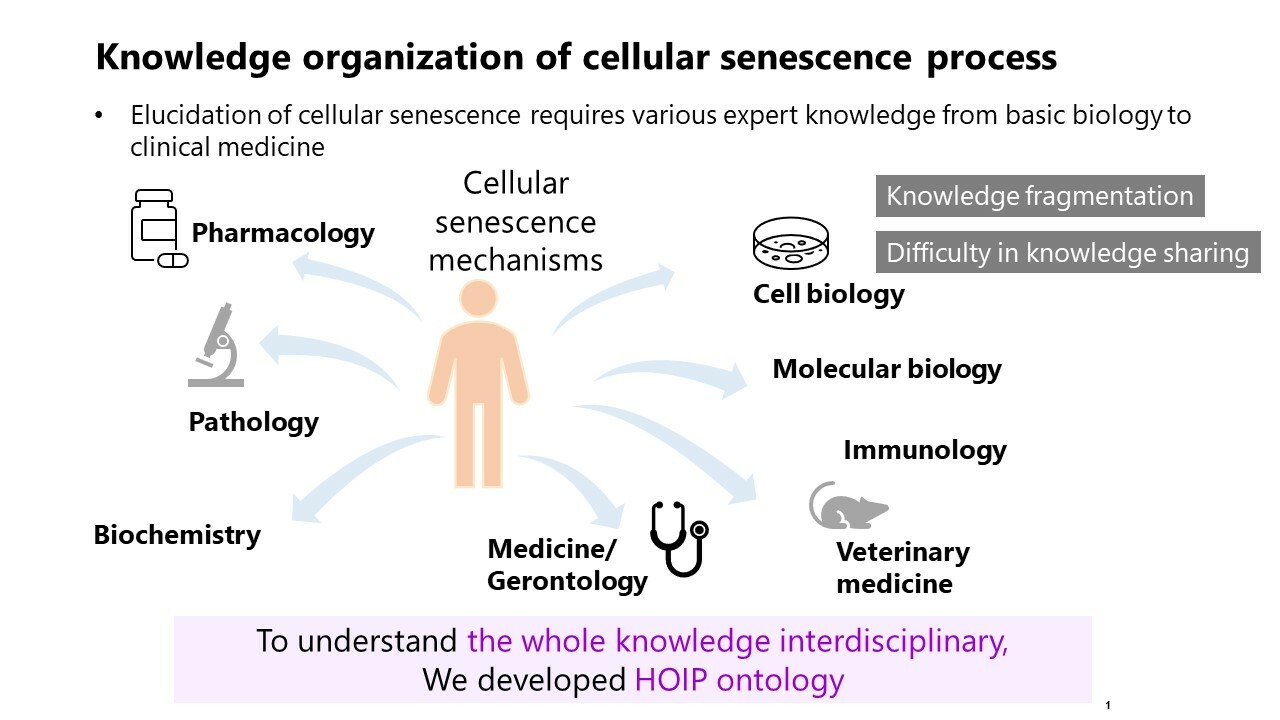
Understanding cellular senescence is crucial for unraveling the complexities of aging and chronic diseases.

Cellular senescence, a state of stable proliferative arrest, plays a significant role in organismal aging and the development of various chronic diseases (Gasek et al.). Despite its importance, the mechanisms underlying cellular senescence are not fully understood. This study aims to integrate and organize existing knowledge to provide a clearer understanding of these mechanisms from a homeostasis imbalance perspective.
What We Did and Why
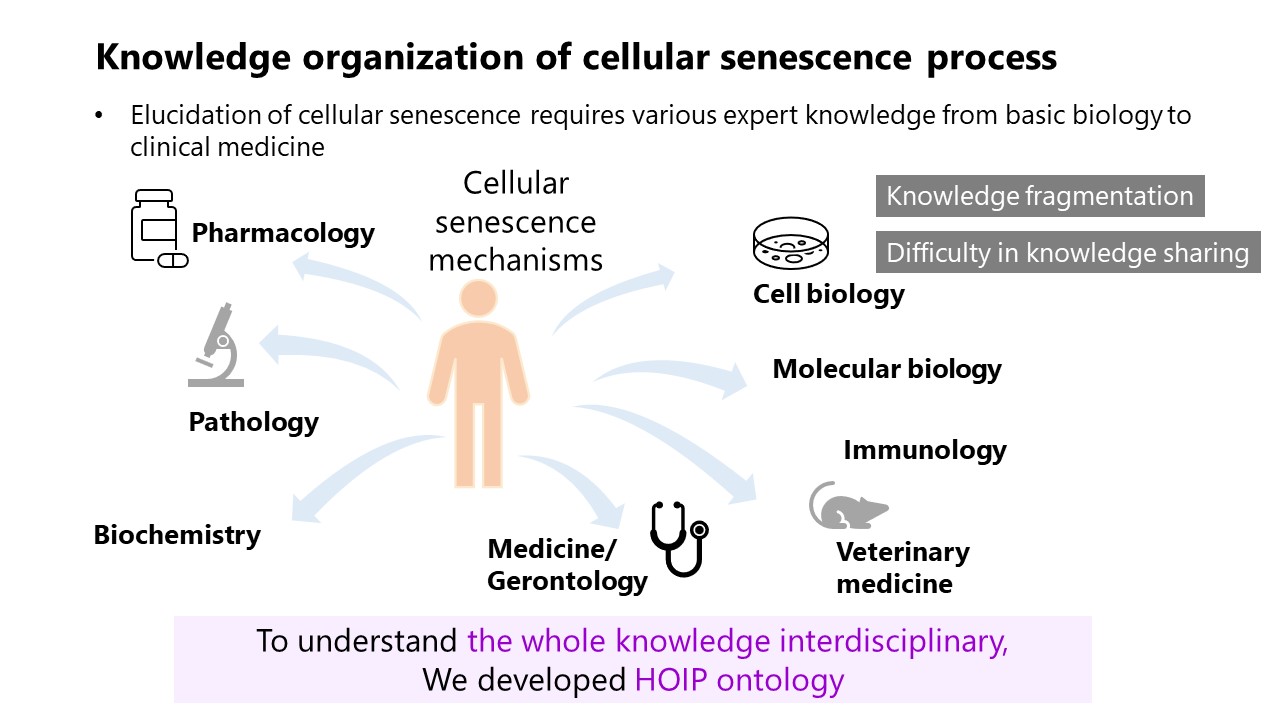
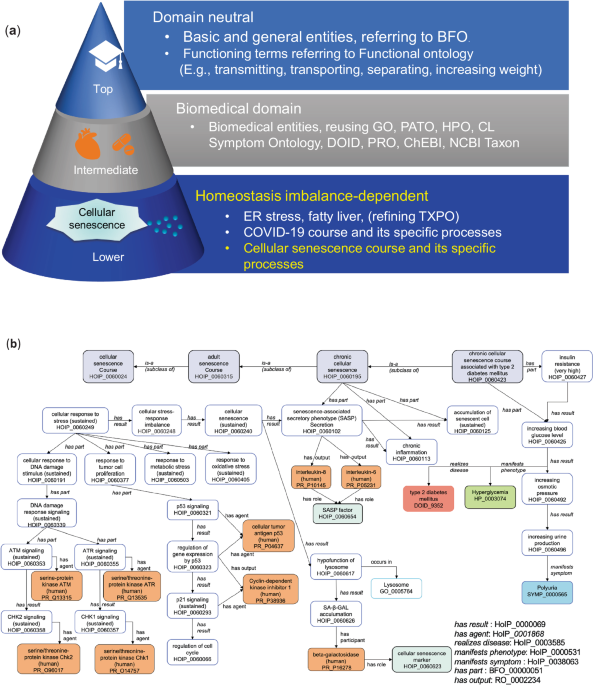
We developed an ontological framework to systematically describe cellular senescence mechanisms. The primary objectives were to:
- Develop an ontological model for cellular senescence (a).
- Organize this knowledge in a computer-readable format (b).
- Model the homeostatic imbalance that occurs during cellular senescence.
We developed the Homeostasis Imbalance Process (HoIP) ontology, integrating terms from existing biomedical ontologies. We then conducted formal verification and content validation to ensure the framework's accuracy and reliability.
What We Found
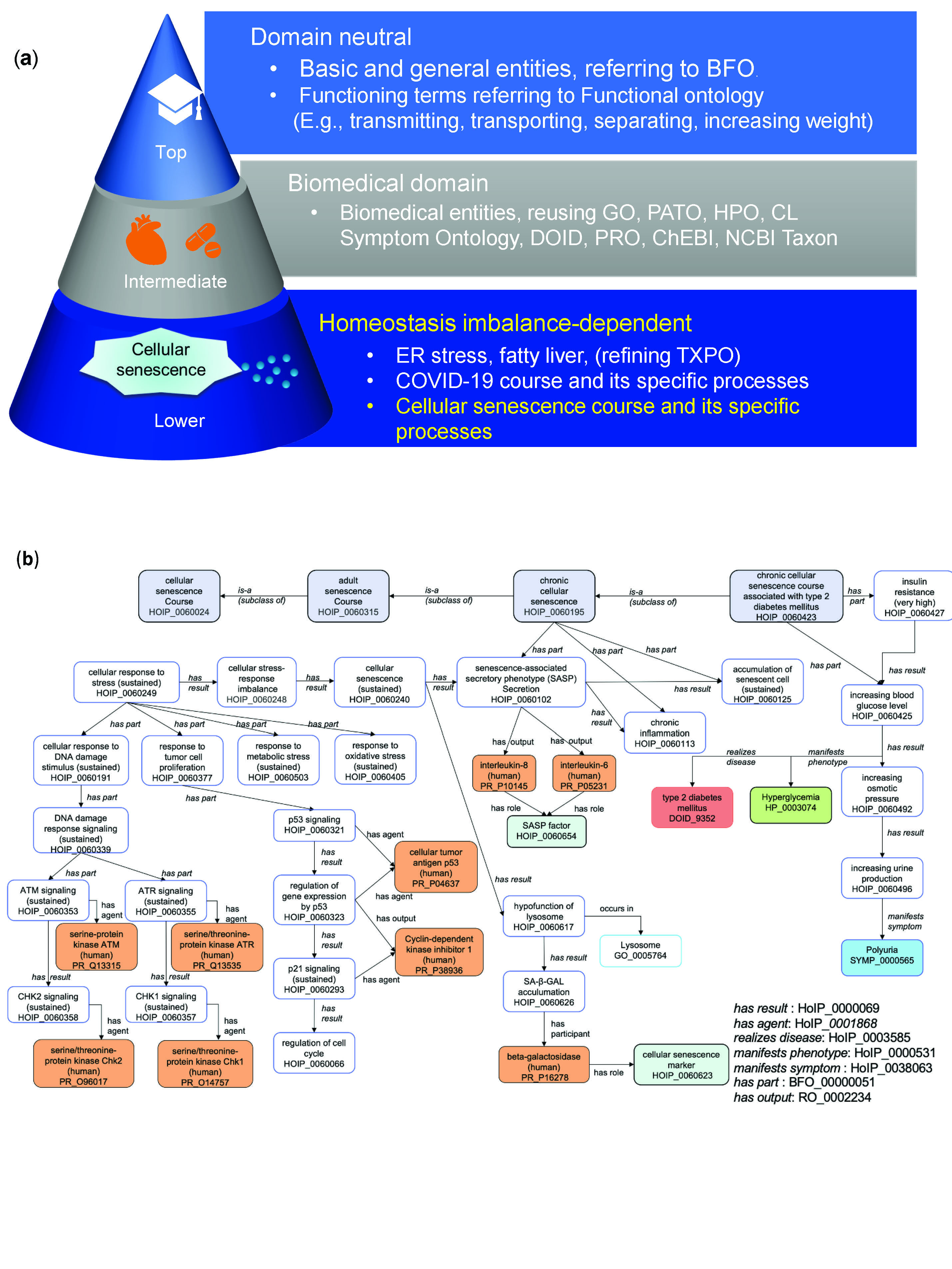
Our research revealed a unified framework for cellular senescence mechanisms based on the viewpoint of homeostasis imbalance. This framework identifies both beneficial and detrimental paths of cellular senescence:
-
Transient Cellular Senescence:
- Types: Embryonic and acute cellular senescence
- Benefits: Tissue remodeling, tumor suppression
-
Persistent Cellular Senescence:
- Type: Chronic cellular senescence
- Detriments: Carcinogenesis due to chronic inflammation via sustained SASP (Senescence-Associated Secretory Phenotype)
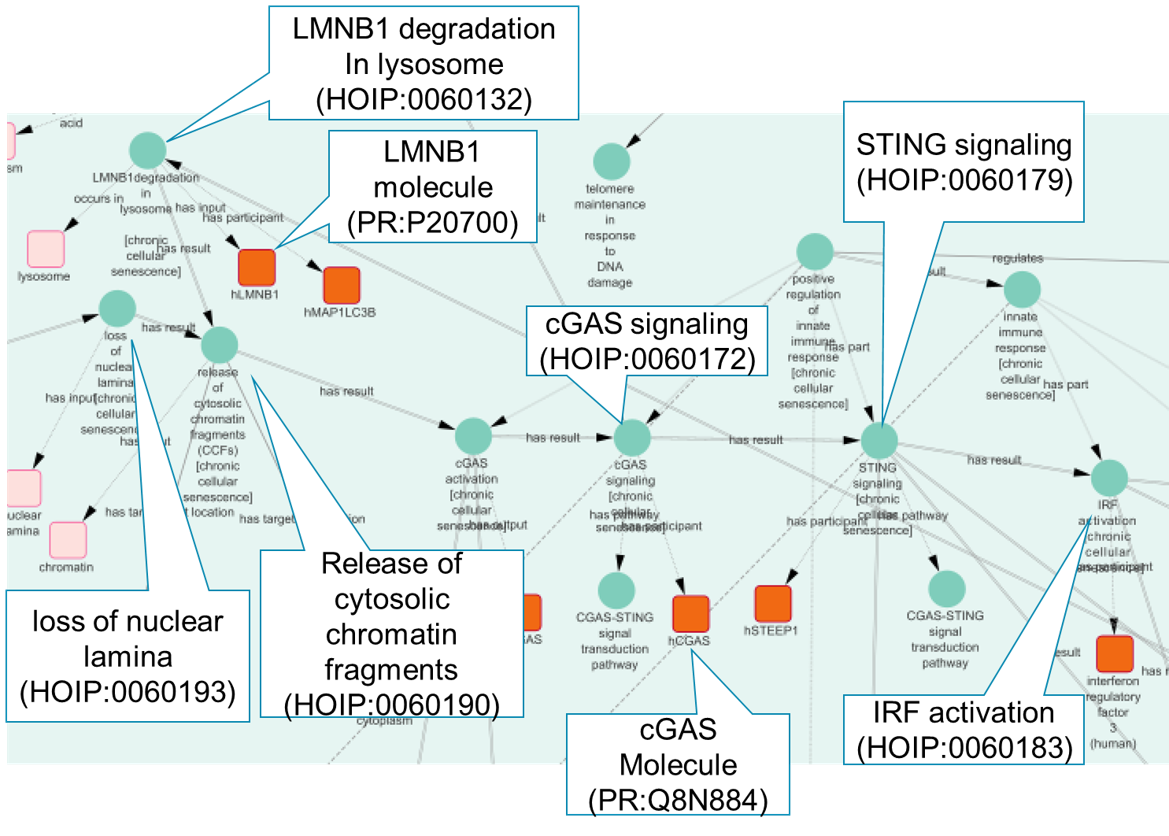
We also applied ontology reasoning to uncover relationships between cellular senescence and diseases, such as type 2 diabetes mellitus and COVID-19. For instance, our analysis suggested that telomere shortening in senescent cells could lead to insulin resistance, contributing to type 2 diabetes. Similarly, SASP factors such as IL-6 and IL-8 were found to be associated with severe COVID-19 outcomes, indicating that cellular senescence mechanisms might play a role in disease severity. Using tools like Cytoscape and NDEx, we visualized these causal networks.
What We Learned, and Future Directions
The study highlights the potential of ontologies to provide a comprehensive and integrated understanding of cellular senescence mechanisms. Our findings suggest that the ontological approach can bridge molecular, cellular, and organ system levels, offering profound insights into disease mechanisms. However, there are limitations, including the manual and time-consuming annotation process and the initial focus on fundamental mechanisms. Future research directions include:
- Expanding descriptions to include more chronic diseases and geriatric syndromes.
- Developing semi-automated annotation processes to improve efficiency.
- Continuously updating the ontology with new research findings.
- Broadening the scope to cover additional areas such as mitochondrial functions and metabolites.
Conclusion
Our ontological framework offers a valuable tool for cellular senescence research, contributing significantly to the understanding of aging and chronic diseases. The framework's potential for future expansions and applications is vast, paving the way for new insights and advancements in the field.
References
-
- Gasek, N. S., Kuchel, G. A., Kirkland, J. L. & Xu, M. Strategies for targeting senescent cells in human disease. Nat. Aging 1, 870–879, https://doi.org/10.1038/s43587-021-00121-8 (2021).
- HoIP ontology-based knowledge exporter Leaves site (https://leaves.riken.jp/)
-
The visualization data are available on the NDEx website (https://www.ndexbio.org/):
Chronic cellular senescence course - Homeostasis imbalance process ontology (https://doi.org/10.18119/N9KS4H)44
Chronic cellular senescence course with type 2 diabetes mellitus- Homeostasis imbalance
process ontology (HOIP) (https://doi.org/10.18119/N9G31J)
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Invertebrate omics
Publishing Model: Open Access
Deadline: May 08, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in