Quantitating Enzyme Activity in Organelles
Published in Bioengineering & Biotechnology
In many cases, a minor population of an enzyme localized within a specific organelle, performs an essential function for the cell. For example, a minor pool of the KDEL receptor in the Golgi is essential to capture ER resident proteins and re-route them to the ER1. While fusing fluorescent proteins to one’s protein of interest provides information on the major population of an enzyme, how might we selectively study the minor pool?
Many small molecule probes are sensitive and specific but report on location-averaged enzymatic activity in cells2. This is because they diffuse rapidly after reaction, blurring spatial information (Fig.1). Genetically encoded reporters do not offer sufficient temporal information and their large sizes often preclude them from being effective substrates3
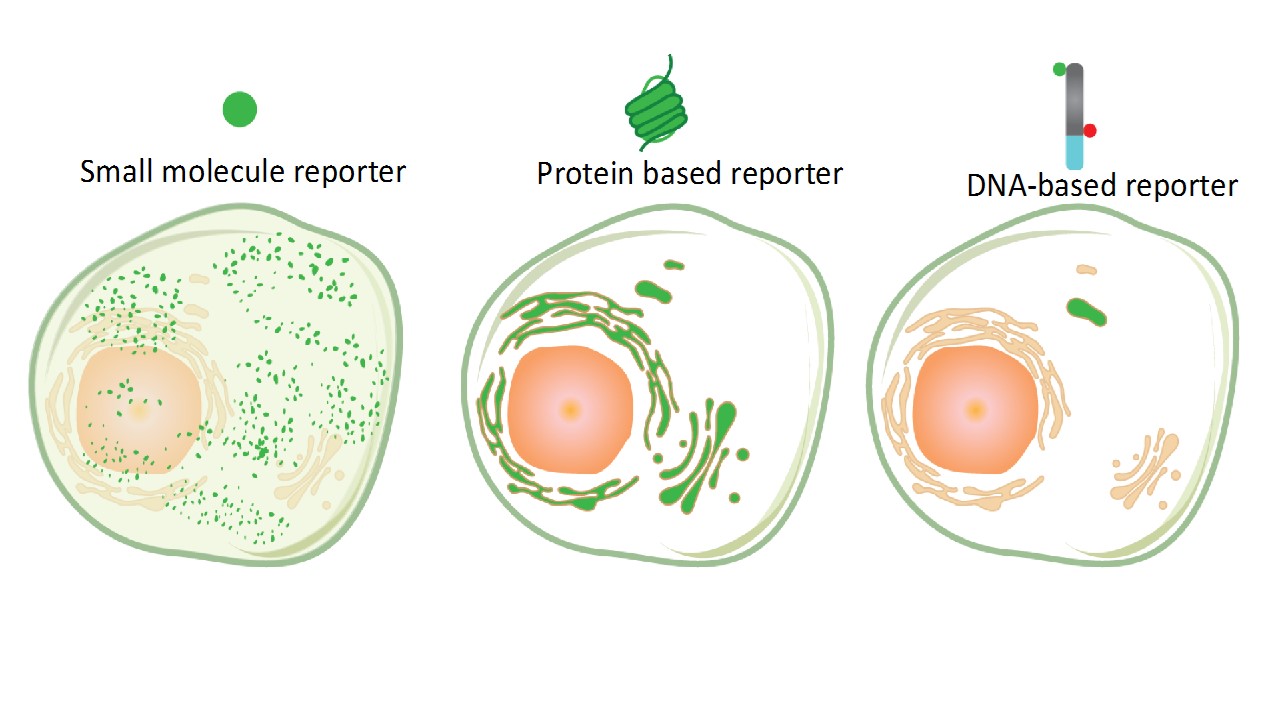
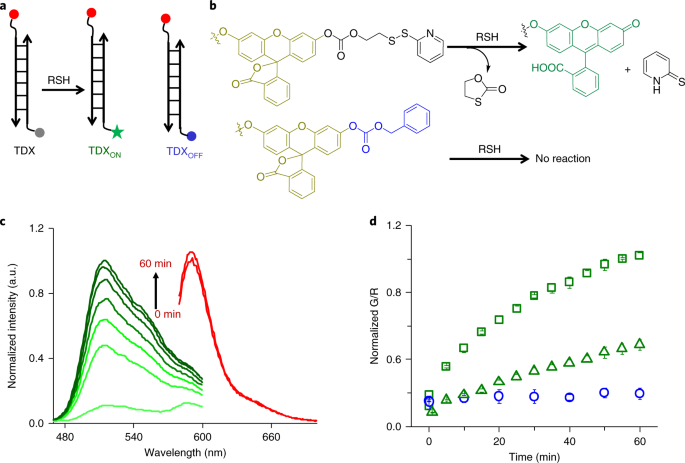
We describe a molecular imaging technology to specifically probe a minor pool of enzyme in organelles without interference from the major population (https://www.nature.com/articles/s41565-019-0365-6). It combines the targetability of genetically encodable probes with the sensitivity and specificity of small molecule detection methods. Our probe is a short DNA-duplex bearing three functionalities i) a small molecule reporter of enzyme activity ii) a reference fluorophore for ratiometric quantitation and (iii) a duplex DNA domain that behaves like an endocytic tracer - localizing in a specific sub-cellular organelle (Fig.1). Post-localization, fluorescence imaging of organelles in live cells as a function of time provides spatial and temporal information on the organelle-resident population of an enzyme.
Here we probe disulfide exchange. This chemistry occurs rampantly throughout the cytosol as well as in many organelles4,5. We showed that we could selectively interrogate disulfide reduction occurring specifically in the endosomes. There has been some debate as to whether endosomal disulfide reduction is mediated by small molecule thiols or whether it is enzyme catalyzed6. Genetic screening using our probe revealed that protein disulfide isomerase (PDI-3) and thioredoxin (TRX-1) were the players that mediated thiol exchange in late endosomes in cells and in vivo. The major populations of these enzymes are in the ER and the cytosol respectively, yet we found a minor pool of each enzyme in the late endosome. We showed that this minor endosomal population was a critical mediator of pathogen infection.
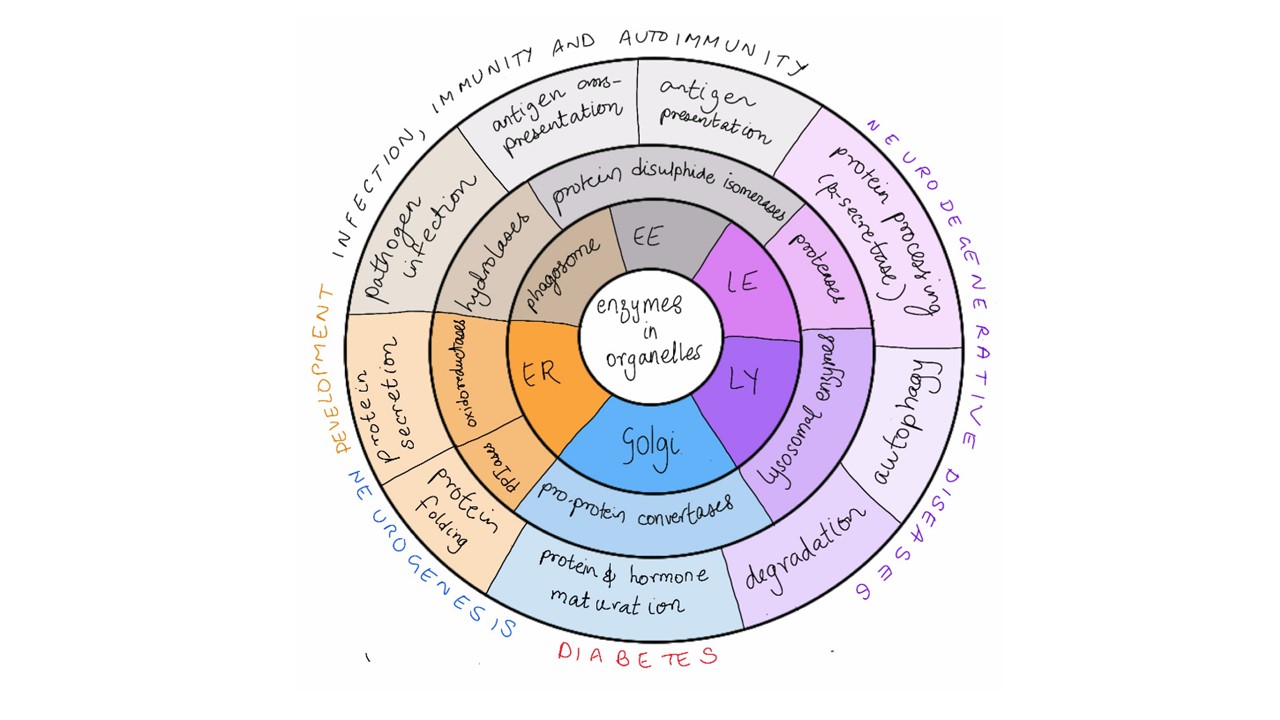
There is a vast array of enzymes in organelles whose dysfunction underlies the development of various diseases (Fig. 2). Detection chemistries for each of these enzymes exist, and now we also have strategies to transport DNA nanodevices to these organelles. This new technology can be expanded to interrogate the activity of many such enzymes.
Lab website: http://krishnanlab.uchicago.edu/
References
1. Dean, N. & Pelham, H. R. Recycling of proteins from the Golgi compartment to the ER in yeast. J. Cell Biol. 111, 369–377 (1990).
2. Liu, H.-W. et al. Recent progresses in small-molecule enzymatic fluorescent probes for cancer imaging. Chem. Soc. Rev. 47, 7140–7180 (2018).
3. Meyer, A. J. & Dick, T. P. Fluorescent protein-based redox probes. Antioxid. Redox Signal. 13, 621–650 (2010).
4. Mesecke, N. et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 (2005).
5. Anfinsen, C. B. Principles that govern the folding of protein chains. Science (80). 181, 223–230 (1973).
6. Yang, J., Chen, H., Vlahov, I. R., Cheng, J.-X. & Low, P. S. Evaluation of disulfide reduction during receptor-mediated endocytosis by using FRET imaging. Proc. Natl. Acad. Sci. USA 103, 13872–13877 (2006).
Follow the Topic
-
Nature Nanotechnology

An interdisciplinary journal that publishes papers of the highest quality and significance in all areas of nanoscience and nanotechnology.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in