RNA structure beyond canonical base pairs guided by evolution
Published in Bioengineering & Biotechnology and Biomedical Research
In addition to messenger RNA (mRNA) that provide the code for proteins, there are other RNAs that exert their function just as RNA, usually referred to as non-coding RNAs (ncRNAs). These ncRNAs have many different functions, from translation (ribosomal RNA, transfer RNAs) to regulation (micro RNAs, riboswitches). Many functional ncRNAs adopt structures specific to their functions. These structures tend to be quite complex and non-local. Much like DNA, RNA structure involves antiparallel double helices of stacked Watson-Crick-Franklin (WCF) base pairs in which nucleotide C pairs with G, and A pairs with T (or U for RNA). Unlike DNA though, RNA helices are usually short, and are connected by loops of unpaired nucleotides. However, these connecting RNA loops are not just unstructured, on the contrary, they are involved in a variety of stabilizing and intricate base pair non-WCF interactions.
The non-WCF base pairs do not form helices, but they are not disordered either. Rather, RNA loops arrange into small structural elements that appear over and over in RNA three-dimensional (3D) structures. These recurrent RNA 3D motifs are the building blocks of intricate 3D configurations that we appreciate in RNA crystal structures. For instance, transfer RNAs acquire a characteristic three-dimensional L-shape resulting from interactions between residues in the loops of two helices.
Because producing RNA crystal structures is still costly, the computational prediction of RNA structure from RNA sequence is useful. There are many algorithms to predict WCF base pairs present in structured RNAs (the secondary structure), and there are also methods to predict 3D motifs given a secondary structure. RNA secondary structure prediction is suboptimal in part due to the omission of the RNA 3D motifs, and the prediction of RNA 3D motifs alone is difficult because motifs are small and do not possess much information content, resulting in many false positives.
In a recent Nature Methods article, “All-at-once RNA folding with 3D motif prediction framed by evolutionary information”, Aayush Karan and Elena Rivas present a computational method that predicts both (secondary structure plus 3D motifs) jointly helping mitigate many of the mentioned difficulties inherent to full structure prediction of RNAs.
The new method, named CaCoFold-R3D, has a number of interesting properties not put together before: it can take into account many different motifs (everything), and it can identify 3D motifs in any non-helical regions (everywhere), all of which are combined in one single prediction (all at once).
But the most important property of CaCoFold-R3D is that it works on alignments. The WCF base pairs (A:U, U:A, C:G and G:C) are all interchangeable in a helix. In consequence, conserved WCF base pairs show a distinctive pattern of co-variation that is well observed in alignments. CaCoFold exploits covariation in alignment to identify helices. Non-WCF base pairs in 3D motifs being of a different kind, are not interchangeable and do not covary. But by predicting these two jointly, 3D motif detection benefits from the significant structural constraints imposed by the covarying helices.
This work originated as an undergraduate research project for Aayush Karan (Harvard 2023), now a Harvard PhD student in Computer Science. Aayush was the first and only first-year that took Elena’s course MCB111 Mathematics in Biology. Aayush produced a prototype incorporating two 3D motif archetypes. The results were so encouraging that Elena decided to do a full integrated implementation, which took a while to get started and to complete. But they persisted, and the combination of their efforts has led to this new method and manuscript.
CaCoFold-R3D is fast, easy to use, and customizable. In addition to jointly predicting WCF base pairs and 3D motifs from alignments of RNAs, it can also be used for de novo discovery of other 3D motifs. Moving forward, the Rivas lab plans to investigate the potential of customizing CaCoFold-R3D to identify RNAs with particular loop motifs interacting with small molecules with therapeutic applications.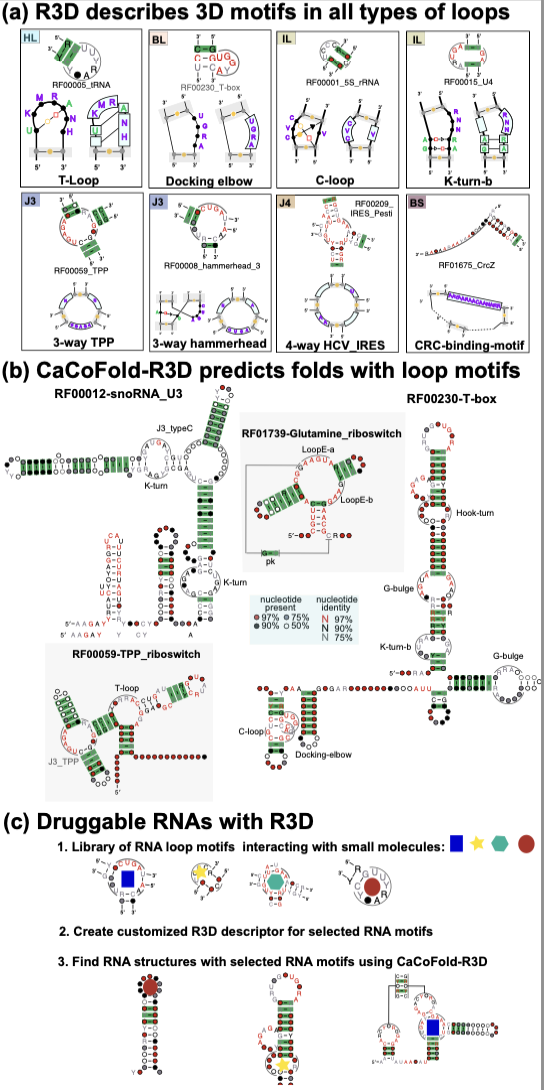





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in