RNAi’s Potential to Tackle the Barber’s Pole Worm – Two Decades-Long Quest for Better Parasite Control
Published in Microbiology and Zoology & Veterinary Science
For small ruminant farmers worldwide, the barber’s pole worm (Haemonchus contortus) is a relentless threat. This blood-feeding nematode causes anaemia, weight loss, and even death in sheep and goats, costing the livestock industry millions annually. For decades, control relied on two mainstays: anthelmintic drugs and vaccines like Barbervax (with cumbersome dosing protocols). But by 2025, a crisis loomed: multiple drug resistance had become widespread, leaving farmers with fewer options to protect their herds.
The application of RNA interference (RNAi), a tool that silences specific genes, to identify essential genes and target candidates for improved control of H. contortus has been proposed for a decade (please see detailed information in our recent paper in Veterinary Research 2025, 56:194). Our team at Zhejiang University had spent over a decade asking a critical question: Can we target the worm’s own biology to stop its infection—without relying on dwindling drug supplies? This question led us to RNAi and three key genes that keep H. contortus alive. Our recent paper shares how this work moved from lab benches to sheep pens—and what it means for the future of parasite control.
The Starting Point: Why Target H. contortus’s “Weak Spots”?
To beat a parasite, you first need to understand its life cycle. H. contortus has two phases: a free-living stage (eggs, L1–L3 larvae) in soil, and a parasitic stage (L4 larvae, adults) in the host’s abomasum (stomach). The critical jump happens when sheep ingest infective L3 larvae: these “dormant” worms activate, shed their protective sheath, moult into L4s, and start feeding on blood. If we could block this jump, we could stop infection before it starts.
Past research had identified genes linked to nematode development, but few had been tested in live animals—a gap we wanted to fill. We focused on three biological processes the worm can’t live without:
Larval activation: The switch from dormant L3 to infective L4.
Moulting: Shedding the cuticle (outer layer) to grow into the next stage.
Haem utilisation: Using iron from the host’s blood to survive (since H. contortus can’t make its own haem).
The Hunt for Target Genes: From Databases to Microscopes
We started by mining genomic and transcriptomic data—public datasets from WormBase ParaSite and our own past work—to find genes tied to these processes.
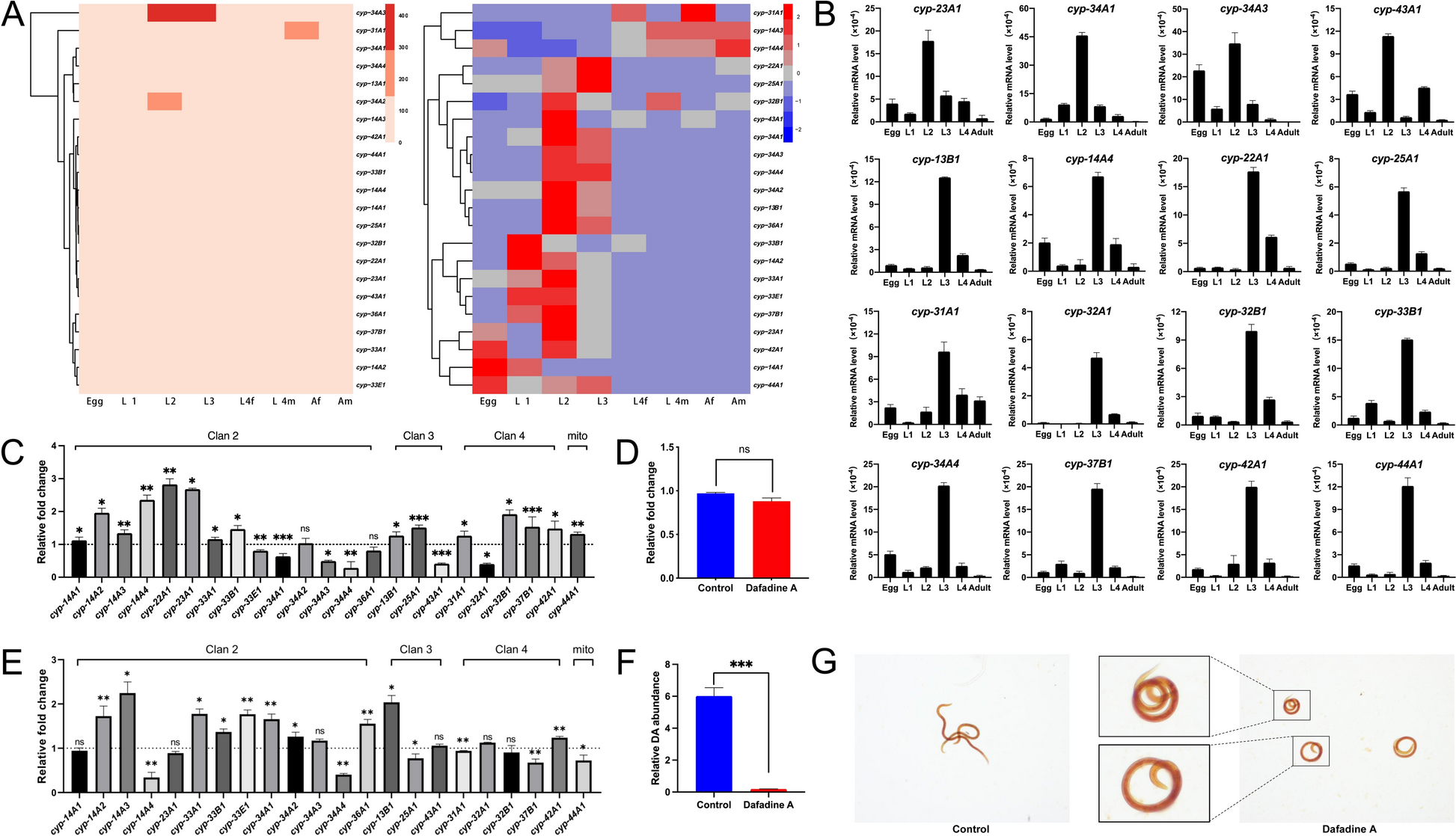
First, we zeroed in on daf-9/cyp-22a1, a gene known to regulate larval development in free-living nematodes like C. elegans. In H. contortus, we found this gene went into overdrive when activated L3 larvae were exposed to sheep serum (mimicking the host’s stomach environment). When we blocked its protein with a chemical inhibitor (dafadine A), the larvae curled up and stopped developing—proof it was essential for the L3-to-L4 transition.
Next, we turned to bli-5, a gene linked to moulting. In C. elegans, bli-5 mutations cause “blister” defects in the cuticle, but no one had studied it in parasitic worms. We discovered bli-5 was highly active in H. contortus’s L3 stage—and that its sequence was only found in nematodes, not in sheep. That was a game-changer: targeting it would avoid harming the host. When we silenced bli-5 in L3 larvae, they developed abnormal bodies (shrinking, swelling) and couldn’t moult properly.
Finally, we looked for genes tied to blood-feeding or haem utilisation. The worm uses a transporter called HRG-1 to take up haem from blood, but HRG-1 is also present in sheep—making it a risky target. Then we found HCON_00083600: a novel gene linked to HRG-1’s function, with no homologs in mammals. It was highly active in blood-feeding L4s and adults, and its protein was concentrated in the worm’s intestine—exactly where haem is absorbed.
The Big Test: From In Vitro Success to Sheep Trials
Lab experiments (measuring larval motility, survival, and gene expression) were promising, but we needed to prove these genes mattered in live hosts. This is where RNAi became our tool of choice.
We used two RNAi methods to silence each gene:
Feeding: For free-living L1–L3 larvae, we fed them E. coli engineered to produce double-stranded RNA (dsRNA) targeting our genes.
Soaking: For infective L3s (xL3s, after sheath removal), we soaked them in small interfering RNA (siRNA) mixed with a delivery reagent to ensure uptake.
Then came the moment of truth: we infected groups of helminth-free Hu sheep with RNAi-treated larvae, alongside control groups (untreated larvae or larvae with non-targeting RNA).
We monitored the sheep for 35 days, measuring:
Faecal egg count (EPG): A marker of adult worm reproduction.
Worm burden: The number of adult worms in the abomasum after necropsy.
The results exceeded our expectations:
When daf-9/cyp-22a1 was silenced: By day 28, sheep had no detectable eggs in their faeces. At necropsy, we found zero adult worms—the silenced larvae never established infection.
When bli-5 was silenced: EPG dropped by ~40% compared to controls, and adult worm numbers fell from ~1,400 to ~800 per sheep. Surviving worms were shorter (a “dumpy” phenotype), meaning they were less able to reproduce.
When HCON_00083600 was silenced: Like daf-9/cyp-22a1, there were no eggs or adult worms in the sheep—proof this novel gene is critical for the worm’s blood-feeding survival.
The Roadblocks: What We Learned (and Still Need to Fix)
This work wasn’t without challenges. RNAi in parasitic nematodes is notoriously tricky: gene silencing efficiency varies, and delivering dsRNA/siRNA to all worm stages (not just larvae) is hard. For example, while we achieved strong silencing in L3s, bli-5 expression bounced back slightly in adult worms—suggesting we need longer-lasting RNAi tools.
We also had to address safety: Could silencing cyp genes (which are part of a large enzyme family) harm the sheep? Because we targeted genes unique to nematodes (like bli-5 and HCON_00083600) or genes with distinct sequences in worms vs. hosts (like daf-9/cyp-22a1), the sheep showed no adverse effects—no weight loss, no changes in bloodwork.
Another lesson: Context matters. In the lab, silencing genes stopped larvae in their tracks, but in live sheep, the immune system also plays a role. We saw that RNAi-treated larvae were less able to evade the sheep’s defences—hinting that combining RNAi with immune boosters could be even more effective.
What’s Next? A New Era for Parasite Control
For us, this paper is just the start. The three genes we identified are now validated targets for H. contortus control, and RNAi has proven it can work in real-world conditions (not just petri dishes). Here’s what we’re working on next:
Better delivery systems: We’re exploring lentivirus-based RNAi to silence genes inside the host, so we don’t have to treat larvae before infection.
Combination therapies: Pairing RNAi with low-dose anthelmintics could reverse drug resistance—a critical need for farmers.
Broadening to other nematodes: Genes like daf-9/cyp-22a1 are conserved in other parasitic worms (e.g., Nippostrongylus brasiliensis), so this work could help control multiple parasites.
A Note of Gratitude
This work wouldn’t have been possible without the farmers who shared their challenges, the students and lab technicians who counted thousands of larvae, and the funding bodies that supported risky, long-term research (including the National Natural Science Foundation of China nos. 32473050, 32202829, and 32172877). Most importantly, it’s a reminder that solving agricultural problems requires bridging basic science and real-world application.
For the research community: We hope this paper inspires more work on RNAi for parasite control—especially in understudied species. For farmers: The end of drug resistance isn’t here yet, but we’re one step closer to tools that protect your herds and the planet.
— Guangxu Ma, and the entire research team
Read the full paper here: https://doi.org/10.1186/s13567-025-01633-6

Follow the Topic
-
Veterinary Research

This is an open access journal that publishes high-quality and novel research and review articles focusing on all aspects of infectious diseases and host-pathogen interactions in animals.
Related Collections
With Collections, you can get published faster and increase your visibility.
Management of infectious diseases in animals: a global view of the current challenges
This Collection seeks to synthesize management strategies used to control infectious diseases in animals and their associated challenges. Each contribution will be framed around a central question (simple to state, but not necessarily to answer): “Given current knowledge, what are the key challenges in managing [disease name], and where are the most promising advances likely to emerge in the next decade?” We encourage papers to adopt a global perspective, enabling comparisons across different contexts (e.g., endemic vs. disease-free regions). Titles will follow a standardized format: “Management of [disease] in [host species]: a global view of current challenges.”
To structure the collection, we will pre-select a set of animal infectious diseases with major impacts on animal and/or human health. These will cover a broad range of host species (livestock, companion animals, fish) and pathogens (viruses, bacteria, parasites, prions), across different regulatory settings (local, national, and international). Internationally-recognized experts will be invited by the guest editorial board to assemble author teams with extensive expertise in managing the disease of interest and representing diverse epidemiological contexts worldwide. Contributors may include academic researchers, risk assessors, or risk managers. While most papers will be commissioned, unsolicited submissions are welcome if the lead author receives prior approval from the editorial board by demonstrating both the significance of the disease and the global expertise of the proposed author team.
The challenges addressed in each paper may relate to logistics, stakeholder engagement, acceptability, technical or technological limitations, regulatory gaps, evidence-based decision-making, or conceptual hurdles. However, these must be presented as specific and well-defined issues. Authors will be asked not to produce general overviews of diseases nor exhaustive reviews but to focus on operational and strategic challenges and to cite only a few carefully chosen references that provide context and support their arguments. This will be reinforced through limits on manuscript length and citation counts. Each paper should outline five to ten key challenges, offering informed perspectives rather than an exhaustive catalogue. Some overlap across papers is expected, as many diseases share common management issues. All submissions will undergo peer review by at least two leading experts to ensure that challenges are significant, clearly presented, and adequately grounded in prior work.
Each article will aim to be accessible and useful not only to researchers, but also to farmers, veterinarians, national veterinary authorities and international organizations (e.g., WOAH, FAO). The goal is to foster dialogue between science and decision-making. The overarching aim of this collection is to present a series of well-defined problems that require solutions, with the hope of inspiring risk assessors and managers to tackle them and strengthen our capacity to manage infectious animal diseases. Success will be achieved if, in the coming decades, many of the challenges described here become focal points for future research, policy discussions, or implementation frameworks. We are excited to play a role in advancing this progress.
Unsolicited submissions are welcome if the lead author receives prior approval from the editorial board by demonstrating both the significance of the disease and the global expertise of the proposed author team.
This Collection supports and amplifies research related to SDG 2, Zero Hunger, SDG 3, Good Health and Well-Being, SDG 12, Responsible Consumption and Production, and SDG 15, Life on Land.
All submissions in this collection undergo the journal’s standard peer review process. Similarly, all manuscripts authored by a Guest Editor(s) will be handled by the Editor-in-Chief. As an open access publication, this journal levies an article processing fee (details here). We recognize that many key stakeholders may not have access to such resources and are committed to supporting participation in this issue wherever resources are a barrier. For more information about what support may be available, please visit OA funding and support, or email OAfundingpolicy@springernature.com or the Editor-in-Chief.
Publishing Model: Open Access
Deadline: Sep 22, 2026
Streptococcus suis, a major swine pathogen with zoonotic impact – advances in epidemiology, pathogenesis, and control
Streptococcus suis remains one of the most significant bacterial pathogens affecting the global swine industry, responsible for substantial economic losses and serious animal welfare concerns. Beyond its impact in pigs, causing mainly meningitis, arthritis, sudden death and endocarditis, S. suis is an important zoonotic agent, causing severe invasive infections such as meningitis, septic shock and other infections in humans, particularly in regions with high levels of pork production and consumption. As such, S. suis exemplifies the necessity of a One Health approach, bridging veterinary medicine, sustainable animal production, and public health. S. suis is also responsible for one third of antibiotic use in post-weaned piglets.
In recent years, remarkable progress has been achieved in understanding S. suis virulence factors, host–pathogen interactions, population diversity, and mechanisms of antimicrobial resistance. Advances in surveillance, diagnostic techniques, immunology, and vaccination strategies are providing new opportunities for disease prevention and improved clinical management. At the same time, changes in pig production systems, global trade, and environmental pressures continue to shape the epidemiology and emergence of S. suis disease worldwide.
Following the 6th International Workshop on Streptococcus suis, that was held in Cambridge, UK in September 2025, this Special Collection brings together state-of-the-art research across the full spectrum of science on this pathogen. Topics include but are not limited to: bacterial genomics and evolution; virulence determinants; host immunity and pathophysiology; vaccines and immunotherapies; diagnostic innovation; epidemiology and transmission dynamics; antimicrobial resistance and stewardship; and clinical aspects in both pigs and humans.
Our aim is to highlight innovative findings, promote interdisciplinary exchange, and foster collaborative strategies to mitigate the burden of S. suis infections. By bringing together contributions from leading experts in the field, this issue will help guide future research priorities and support the development of effective disease control measures benefiting animal and human health alike.
We warmly invite submissions of original research articles, short communications, and comprehensive reviews.
Additional information:
The collection features contributions presented at the International Workshop and invites complementary studies that expand upon its themes.
This Collection supports and amplifies research related to SDG 3, Good Health and Well-Being and SDG 12, Responsible Consumption and Production.
All submissions in this collection undergo the journal’s standard peer review process. Similarly, all manuscripts authored by a Guest Editor(s) will be handled by the Editor-in-Chief. As an open access publication, this journal levies an article processing fee (details here). We recognize that many key stakeholders may not have access to such resources and are committed to supporting participation in this issue wherever resources are a barrier. For more information about what support may be available, please visit OA funding and support, or email OAfundingpolicy@springernature.com or the Editor-in-Chief.
Publishing Model: Open Access
Deadline: Aug 20, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in