Roles of bacteriophages, plasmids and CRISPR immunity in microbial community dynamics revealed using time-series integrated meta-omics
Published in Microbiology
The 2020 Nobel Prize in Chemistry was awarded to Emmanuelle Charpentier and Jennifer Doudna for the discovery of the genetic editing tool, CRISPR/Cas9. While genetic editing has been possible prior to this discovery, CRISPR/Cas9 achieves a much higher precision. It is widely regarded as one of the most important discoveries of the century, with profound impact on medicine and biotechnology.
The CRISPR/Cas9 system is a modified version of a naturally occurring bacterial immune mechanism against (bacterio)phages, i.e. viruses infecting bacteria. There are many different types of CRISPR-Cas systems (other than CRISPR/Cas9) that protect bacteria from phage infections. Phages are thought to be some of the most abundant and diverse biological entities on the planet. Hence, they are believed to strongly influence microbial community structures and functions. Therefore, our goal was to understand the influence of phages within a naturally occurring microbial ecosystem (Video 1).

Video 1: Foaming activated sludge islets floating on the surface of a biological wastewater treatment plant and how the density evolves over one and a half year.
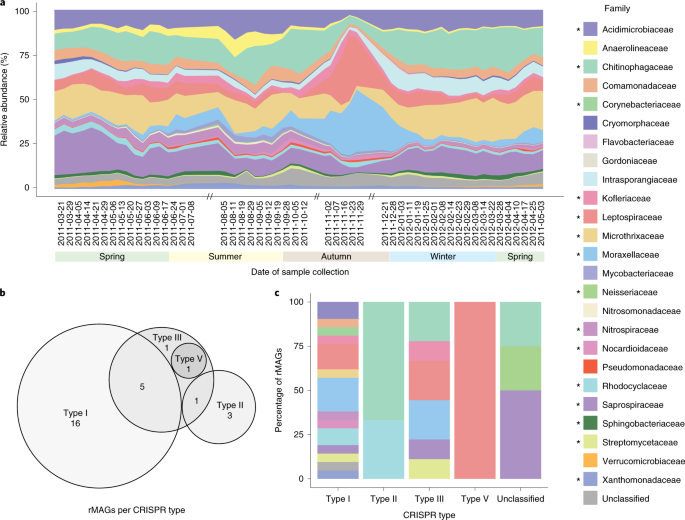
The generation of an unprecedented meta-omics data set provided us a great opportunity to study the effect of phages within a naturally occurring microbial community of a biological wastewater treatment plant. We identified 31 bacterial metagenome-assembled genomes (MAGs) encoding complete CRISPR-Cas systems, while we used the spacer information from the CRISPR locus to identify the sequences these CRISPR systems were targeting. We assumed most of these targeted sequences to be invasive mobile genetic elements (iMGEs), out of which a large majority would be phages. However, to our surprise, not only did we find that plasmid sequences were 16 times more abundant than phages within the community, but that they were also targeted five times more frequently than phages (Video 2). Although CRISPR-Cas systems are known to target other iMGEs, such as plasmids, it was previously thought that phages were targeted predominantly. To the best of our knowledge, we were first to show such consistent prevalence of plasmids being targeted by CRISPR-Cas systems.
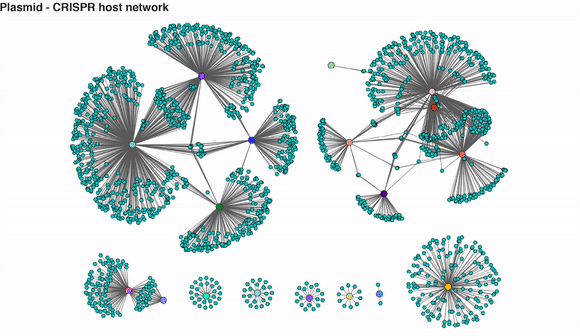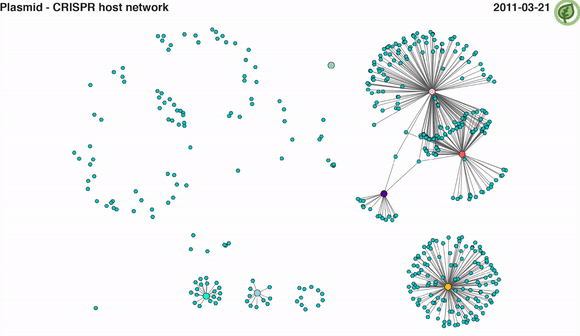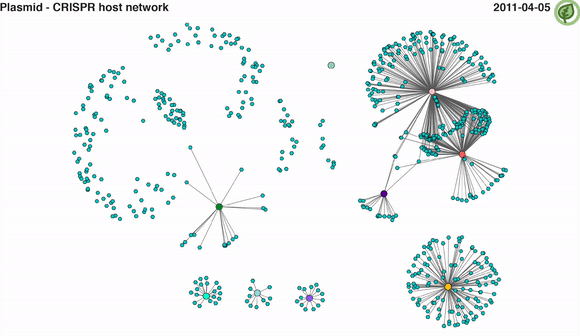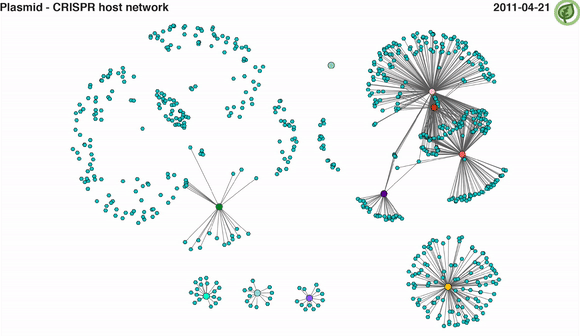
Video 2: CRISPR-mediated plasmid-host interaction networks over time. Turquoise nodes represent plasmid sequences, while larger coloured nodes represent bacterial MAGs. Edges between nodes represent spacer-protospacer links/matches. Edges in a given time point appear when both plasmid sequence and its linked MAG are present.
In addition, we estimated the relative importance of plasmids and phages on the dominant microbial population dynamics through linear modelling. The models emphasised the impact of plasmids on the dynamics of the dominant population (Figure 1). Moreover, our results suggest bacterial CRISPR systems were selective towards retaining plasmids encoding genes of beneficial functions, for example antimicrobial resistance genes.
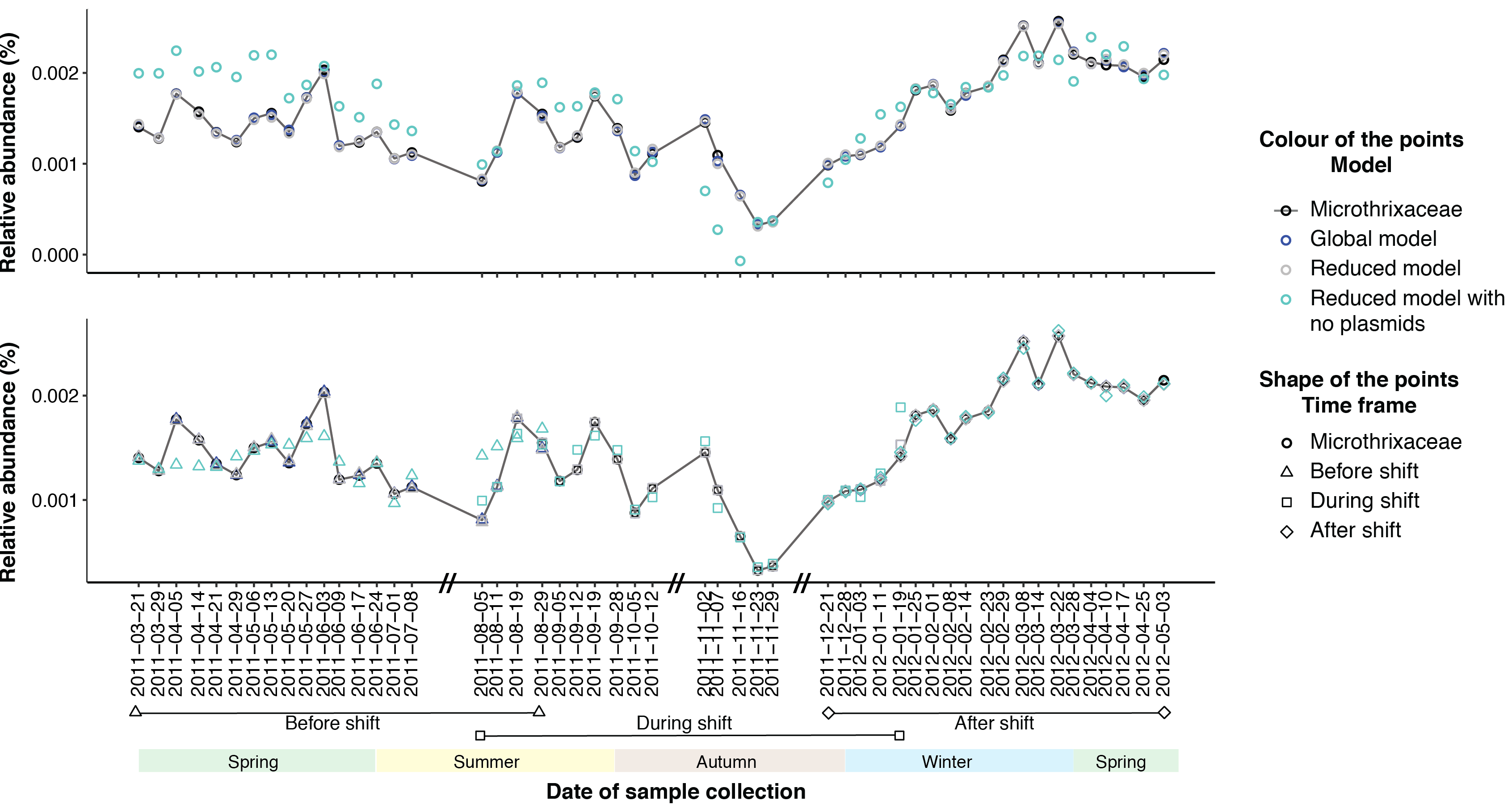
Figure 1: Data fit to linear models that were performed for the entire time series (top panel) and for shorter time intervals (bottom panel). The models predict the abundance of Microthrixaceae.
We then explored the CRISPR-Cas systems of several key populations to resolve iMGE-host interactions at the population level. Using our meta-omics data set, we observed that the dominant population of the community (Microthrix parvicella) had an active CRISPR locus, with integration and deletion of CRISPR spacers (Figure 2), while the cas genes were expressed both on the RNA and protein level. A similar observation on another population (Leptospira biflexa) demonstrated more CRISPR spacer integration and deletion events compared to the dominant population, suggesting that some populations use their CRISPR systems more than others.
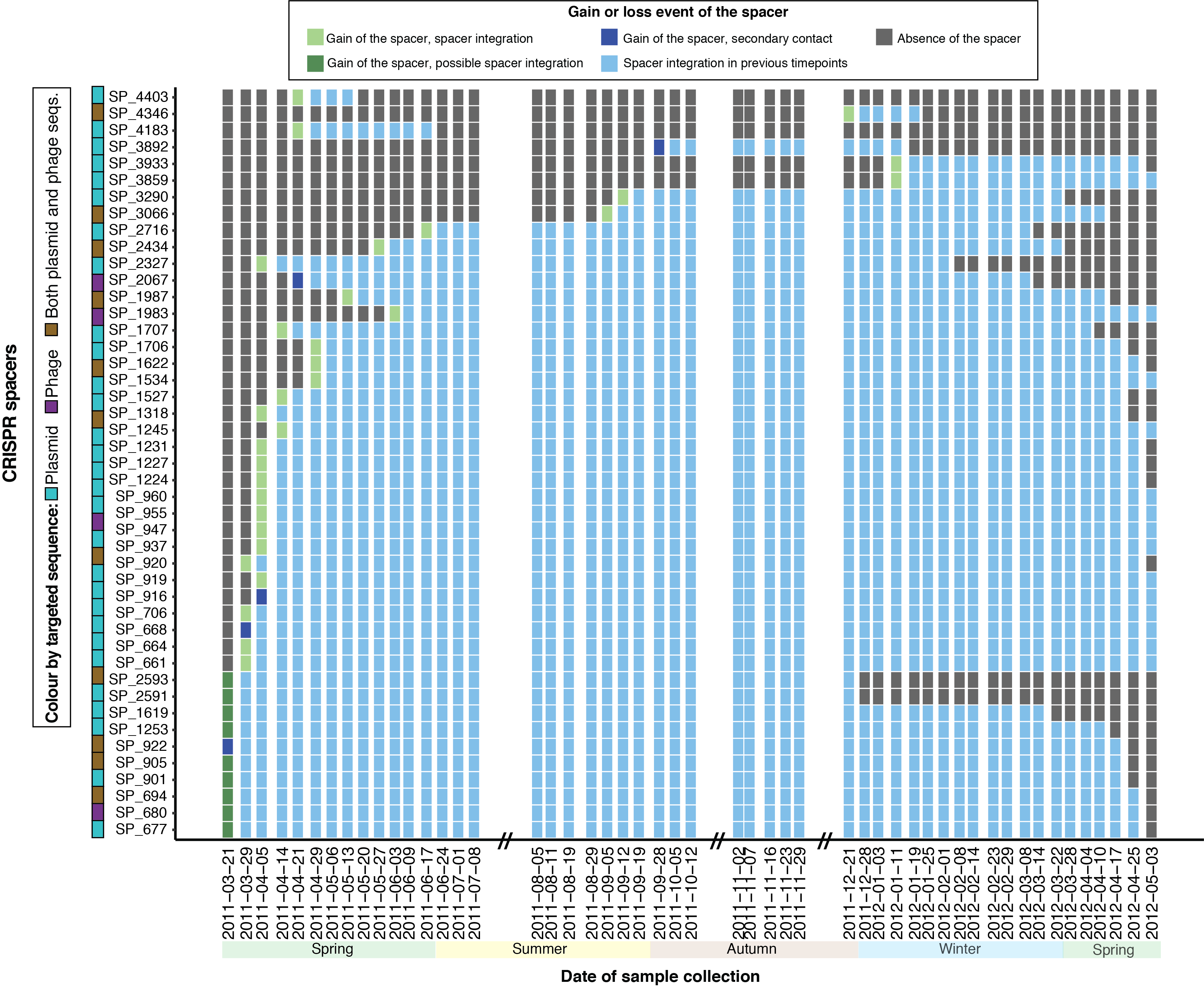
Figure 2: CRISPR spacers of Microthrix parvicella are active (i.e., are gained or lost) over the time series.
Our findings capture the interactions that play key roles in promoting adaptation, diversity and resilience within microbiomes. Understanding the factors that impact microbial populations is crucial as it can help to both devise measures against deleterious species and to predict how the community will evolve over time. Based on our observations, iMGEs and CRISPR-based interactions are important factors when studying and modelling microbial communities. The knowledge from these models can be used to engineer wastewater treatment plants or other important microbial communities, such as those within the human gut. Interestingly, direct engineering of microbial consortia may now be achieved using state-of-the-art CRISPR-Cas technologies, bypassing the need for isolated cultures, as suggested by Rubin et al. In summary, the combination of enhanced knowledge and expansion of the gene editing toolkit could bring about significant outcomes in biotech and biomedicine.
On a separate note, this is the sixth publication stemming from this project and sample set, since its inception, with the first one being published in 2014 and the last one (fifth) in 2020. As it stands, this data set covers 67 biological samples that yielded 121 sequencing data sets (isolate genomics, metagenomics, and metatranscriptomics; NCBI Bioproject PRJNA230567) and 53 metaproteomic data sets (PRIDE PXD013655). Given the scarcity of meta-omics longitudinal in situ data, we hope that other researchers will use this rich publicly available data set to further explore other aspects of this system. We believe that continued collection efforts as well as longitudinal sampling and analysis are crucial to better understand microbial ecology in environmental or host-borne samples.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in