Screening Host Defense Peptide Libraries for Antibiofilm and/or Immunomodulatory Activities
Published in Protocols & Methods
In recent years, we have seen an increasing interest in the research field of antimicrobial HDPs and their synthetic analogs /or mimetics, which is perhaps due to the rise in their appreciation of multimodal functionalities and the acknowledgement of their therapeutic potentials. This positive growing interest could indeed help speed up the possibility of finding new antiinfective or anti-inflammatory agents based on HDPs.
We understand that, like other drugs, the development of HDPs as therapeutics is not straightforward; the process can be tedious and complicated. We also know that, unlike other drugs, the knowledge we have about the multifaceted functionality of HDPs, such as their antibiofilm or immunomodulatory properties and the sequence constraints that define strengths related to their bioactivities, is very minimal. Extensive work needs to be done to fully reveal the structural activity relationship of these HDPs, to enrich our knowledge of their diversity, sequence limitations, mechanism of action etc. Our aim of this Nature Protocols' article was to address this issue (to some extent) and describe highly reproducible approaches that can be used regularly to assess hundreds of HDPs biological activities, including biofilm inhibition and immunomodulatory functions, through integrating the SPOT-peptide array technology with high-throughput screening assays.
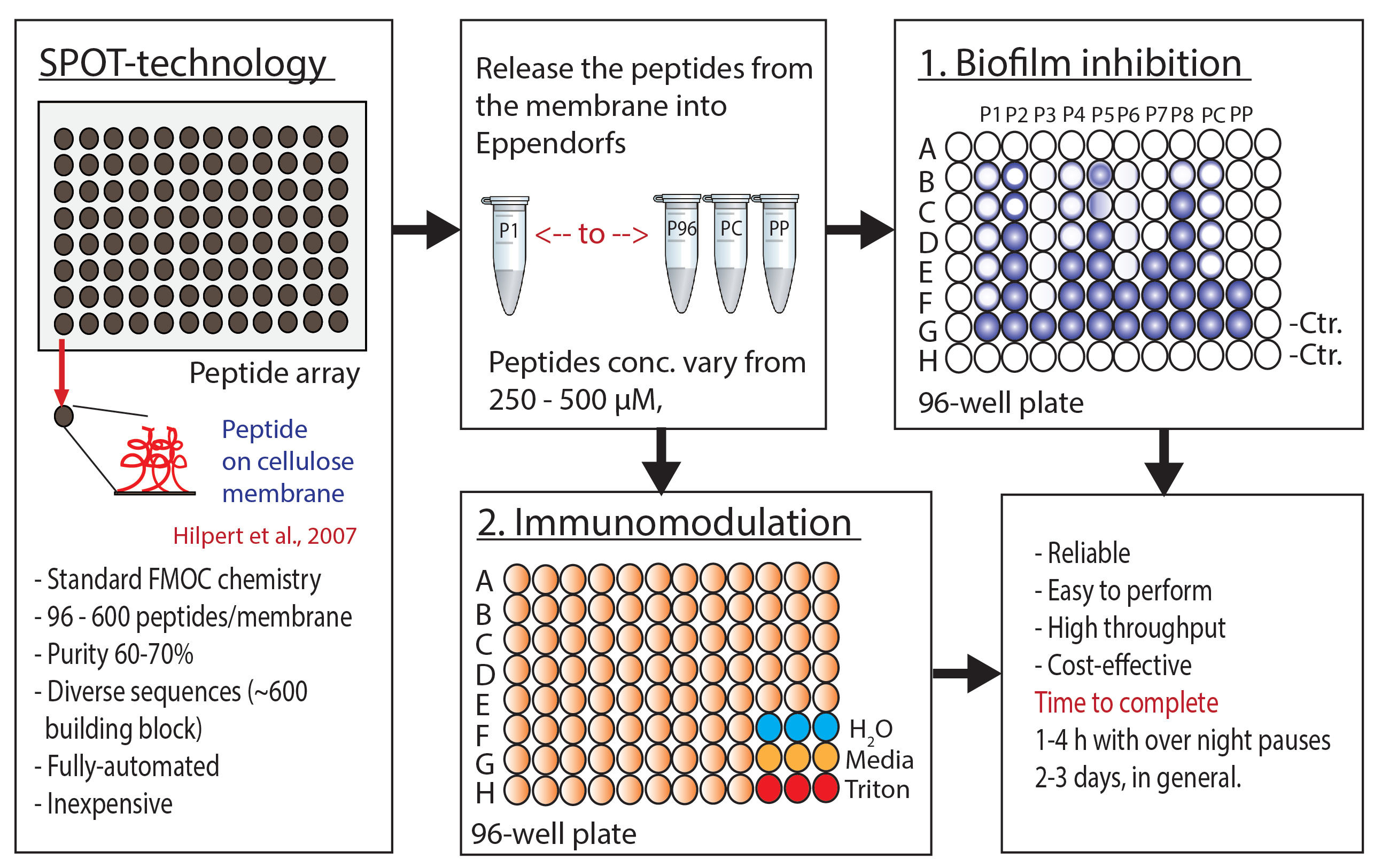
A simple diagram of the protocol.
In our lab's earlier Nature Protocols' Protocol by Hilpert et al., 2007. a systemic approach to synthesizing arrays of peptides on cellulose membranes using SPOT-technology was described – a method pioneered by Ronald Frank and his co-workers in 1990. The strategy offers a synthesis of a large number of peptides in small concentrations and at meagre cost (~1% of the cost/peptide) relative to the conventional Solid Phase Peptide Synthesis (SPPS). However, the method still follows the standard chemistry on SPPS and can utilize more than 600 different building blocks that dramatically enhance the peptides' structural diversity.
In today's follow-up Nature Protocols' Protocol, we extend the previous protocol's mission and describe in details screening methods accessible for confined amounts of samples to assess the antibiofilm and immunomodulatory activities of those SPOT-array synthesized HDPs. We divided the protocol into two main sections. We showed the antibiofilm activity assessment, in a term of biofilm inhibition assay using the crystal violet staining in the first part. Subsequently, a procedure of testing the immunomodulatory activity of the same SPOT-peptides against peripheral blood mononuclear cells is described in the second part. The results of such screening experiments can indeed help us to set the foundation needed for optimizing and designing novel peptide sequences that may become feasible antiinfective and/or anti-inflammatory drug candidates. We applied and still applying the protocols in several studies, and we have been very successful in obtaining potential antibiofilm and immunomodulatory peptide candidates from the screenings.
This protocol's advantages are that it is highly reliable, rapid, easy to perform, and, most importantly, a cost-effective approach for screening peptide arrays. We hope that, collectively, the previous and current Nature Protocols' articles can help researchers in many different ways to find synthetic peptides with promising biological activities.
Article DOI: 10.1038/s41596-021-00500-w. Link: https://rdcu.be/cioLO
Written by Dr Hashem Etayash on behalf of coauthors Dr Evan F. Haney and Prof Robert E.W. Hancock
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in