SeqCode: Towards a unifying system for the naming of cultured and uncultured prokaryotes
Published in Microbiology

Stirring the possum
It has been recognized for decade that the art of scientific naming is rigid and largely unmodernized in the field of microbiology. The exclusion of uncultured prokaryotes from the official nomenclature system has led to chaos and confusion in scientific communication about the microbial world. Most prokaryotes cannot be easily grown in the laboratory and distributed as pure cultures, a requirement for naming under the International Code of Nomenclature for Prokaryotes (ICNP). As-yet-uncultivated taxa account for ~85% of the phylogenetic diversity of the prokaryotes1, thus a substantial portion of the tree of life cannot be officially named. This has stirred numerous debates and eventually resulted in proposal of two paths under which nomenclature of uncultured taxa could be modernized. The so-called ‘Plan A’ envisioned the incorporation of genomic sequences as nomenclature types into the ICNP framework but unfortunately failed following the rejection of this proposal by the International Committee on Systematics of Prokaryotes (ICSP) in March 20202. This triggered ‘Plan B’ under which a new nomenclature code has been developed – the Code of Nomenclature of Prokaryotes Described from Sequence Data, colloquially known as the SeqCode.
Changes are inevitable
So, what can the SeqCode offer microbiologists that ICNP cannot?
First, it allows naming microorganisms based on their genomic sequences rather than pure cultures alone, thus embracing the proposal of Whitman3. The sequence acts as a nomenclature type or simply ‘type’, a concept that is central to nomenclature codes in biology. Type is a reference or comparison standard, which serves as an anchor for a given taxonomic name. Types are usually not typical since the discoverer of a new taxon is usually unfamiliar with the full range of variation within the taxon4. Therefore, their major role is to be a reference of a name and aid in application of that name to newly discovered taxa.
Second, SeqCode allows registration and validation of names in a single centralised web portal, the SeqCode Registry (https://seqco.de/). The registry facilitates the process of name validation and indexing, thus offering an easy tracking system for names, their types, nomenclature status, associated literature and alternating taxonomic opinions. Validation of names in the registry is performed via both automatic and manual checks based on inbuilt standards of genomic sequence quality, which offers greater standardisation across the nomenclature framework.
Third, the SeqCode aims at universality and inclusiveness as it is not only recognising names validly published under the ICNP but it also allows researchers to name their taxa regardless of cultivation status. These principles of universality and inclusiveness are at the core of enabling integration of the existing and new names into a single taxonomic framework.
Overall, these changes are inevitable, necessary, as well as long overdue. We are proud to report that the idea of the SeqCode and its accompanying registry has been strongly supported by the community following a series of international workshops, online debates and conference discussions (See https://www.isme-microbes.org/seqcode-initiative).
Three paths of naming under the SeqCode
There is no need to convince a scientist that the success of any new development depends on its implementation. We took this seriously and are developing three different paths that will allow the registration and validation of taxonomic names.
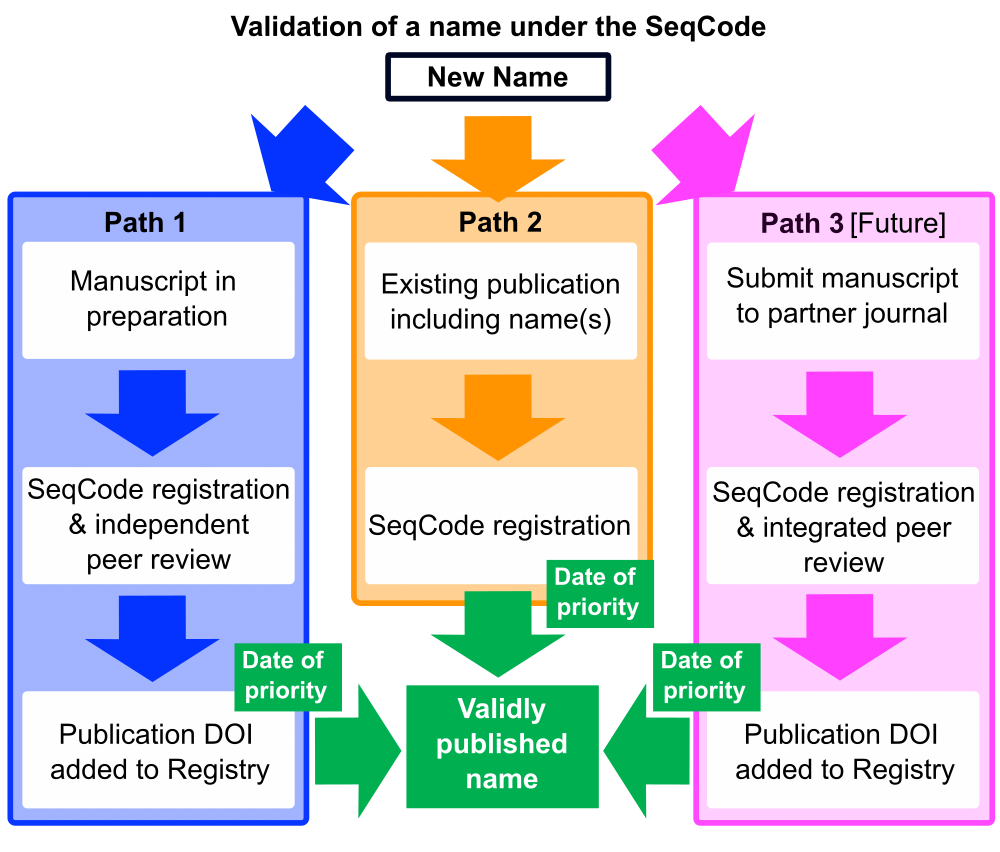
Path 1 is currently recommended and involves pre-registration of taxonomic names and metadata before publication, thus enabling automated checks and curators to guide researchers during the naming process. Path 2 enables registration of existing names including Candidatus names (provisional names that are proposed for the uncultured taxa outside the official nomenclature). This path is deliberately flexible so that it can accommodate such names and emend them if necessary upon validation with all crucial information being captured and available within the registry. Path 3 is also anticipated, in which the publication and registration would occur simultaneously.
Name formation: meaningful and meaningless
Modernization of the nomenclature system does not mean erasing the past. By contrast, it builds on prior rules and principles, thus allowing for an easier transition. Although the SeqCode differs in some essential aspects from the ICNP, its core remains very similar to the existing framework, including usage of Latin as the official nomenclature language. One of the main principles of both the ICNP and SeqCode states that “the primary purpose of giving a name to a taxon is to supply a means of referring to it.” Below we would like to clarify and expand on this statement.
In 1958 William Kent expressed with elegance and brevity what can constitute the conflict between the theory and practice of scientific naming in one phrase: “Words are restrictive only when our attitudes are narrow”5. From the perspective of a taxonomist, one can embrace its sense in which naming can be viewed as a framing to which novelty can be added as our knowledge expands. Restricting this expansion would mean refusing scientific progress. Let’s be less philosophical now and get this idea down to Earth. Some people may truly believe that taxonomic names have meanings but let’s ask ourselves: “How can this be compatible with the practice of taxonomy?”. The answer is “it cannot”. To explore this idea, Kent5 compared the practice of scientific naming of two famous namegivers – French chemists Antoine Lavoisier and English physicist and chemist Michael Faraday:
“In a way, Lavoisier and Faraday had opposite attitudes towards naming. Lavoisier said names should be truly significant of things, and tried to build essential meanings into the vocabulary of chemistry. Faraday, on the other hand, was at times so anxious to avoid names carrying built-in false meanings, that he considered using nearly meaningless terms. Yet in spite of this difference, both Lavoisier and Faraday took names too seriously…” (Kent, 1958)
We invite you to read more on this in Kent’s paper but in an essence – taxonomic names cannot have meanings fixed to them as taxonomy is a dynamic field and therefore meanings evolve and change with time as our knowledge develops. The only meaning that a taxonomic name truly possess is associated with its category in a hierarchical taxonomy and the way it is applied to that category. If we would consider the meaning of a name as a precise label reflecting species properties or history, we should then modify the name each time a new strain with distinct properties is added to it. Obviously, this is neither desirable nor practical. Nomenclature calls for stability of names and changes are only permitted when they are taxonomically well justified and necessary. We believe that building the nomenclature framework based on genome sequences as types will minimize the need of changes and eventually lead to the alignment between nomenclature and taxonomic classifications in the age of the big sequence data.
Concluding remarks
I would like to thank all members of The SeqCode Organizing Committee for the wonderful and productive time that we had working towards the development of a new nomenclature code and registration system. Our mission here is to serve the community of microbiologists from different disciplines, countries and institutions to facilitate naming practice for all prokaryotes and specifically to stabilize the nomenclature of uncultured taxa. We greatly value community feedback on this initiative and invite everyone to participate in its further development and evaluation. I believe that this initiative will not only facilitate scientific communication around microbial diversity but also serve as an essential step towards a unified nomenclature framework for all prokaryotes.
References
- Nayfach S, Roux S, Seshadri R, Udwary D, Varghese N, Schulz F, Wu D, et al. A genomic catalog of Earth's microbiomes. Nat Biotechnol 2021;39:499-509.
- Sutcliffe IC, Dijkshoorn L, Whitman WB, ICSP Executive Board. Minutes of the International Committee on Systematics of Prokaryotes online discussion on the proposed use of gene sequences as type for naming of prokaryotes, and outcome of vote. Int J Syst Evol Microbiol. 2020;70:4416–7.
- Whitman WB. Modest proposals to expand the type material for naming of prokaryotes. Int J Syst Evol Microbiol. 2016;66:2108–12.
- Witteveen, J., 2015. Naming and contingency: the type method of biological taxonomy. Biology & Philosophy, 30(4), pp.569-586.
- Kent, W., 1958. Scientific naming. Philosophy of Science, 25(3), pp.185-193.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in