
The journey of a virus in a host cell is a complex and dynamic process involving multiple infection pathways and steps, dissociation of viral components, and complex interactions with cellular structures. Only a comprehensive understanding of the dynamics and mechanisms of viral infection may help in the prevention and treatment of viral diseases and in the development of antiviral drugs. The quantum dot (QD)-based single-virus tracking (QSVT) technique is an imaging approach that uses QDs as fluorescent tags to label different viral components and fluorescence microscopy to investigate the infection process of single viruses and the dynamic relationship between viruses and cellular components. In QSVT experiments, the behavior of a single virus or viral component labeled with QDs can be monitored within a host cell for long term and in real time. After reconstructing the movement trajectories of individual viruses, the associated dynamic information can be extracted meticulously to reveal the infection mechanism of viruses. Therefore, QSVT is an exquisitely selective and powerful approach to investigate how viruses are internalized into host cells dynamically to release their genome for viral replication and assembly that ensure the completion of viral life cycles. The key steps in performing QSVT experiments are shown in Figure 1.
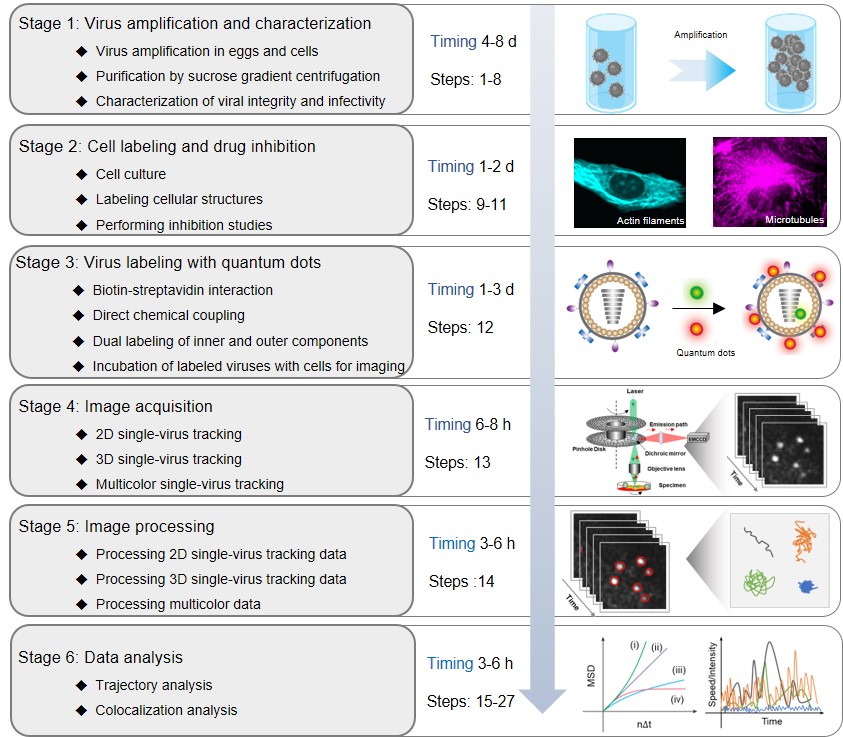
To obtain labeled virus samples for imaging, protocols for the propagation, purification and characterization of virus depend strongly on the virus type (Stage 1). Next, cell structure labeling and drug inhibition assays can be used to facilitate the exploration of the dynamic interactions between virus and cellular structures and the steps of the infection process (Stage 2). Labeling viral components with QDs is crucial for QSVT. The appropriate labeling method needs to be carefully chosen to avoid affecting the viral infectivity. . (Stage 3). Then, labeled viruses can be individually tracked with two-dimensional, three-dimensional or multicolor QSVT techniques by spinning-disk confocal microscopy to obtain time-series images (Stage 4). After that, image processing is required to convert the original stack of fluorescent images into trajectories (Stage 5). Finally, based on the time-dependent plot of speed or intensity, mean-squared displacement and colocalization, the trajectories are statistically analyzed to uncover the underlying mechanisms of virus infection (Stage 6).
Compared to strategies using organic dyes and FPs as tags for SVT, QSVT has significant advantages in the following aspects: i) the extremely high brightness and excellent photostability of QDs facilitate high-contrast and long-time span imaging of individual viruses from milliseconds to hours, and allow for the localization of individual viruses with nanometer-level precision. ii) the color-tunable emission with a narrow full width at half-maximum makes QDs an excellent label for synchronous multi-component labeling of viruses with single virus sensitivity. iii) the abundance of surface ligands on QDs facilitates the labeling of different components of the virus by various labeling strategies. By further combining QDs labeling strategies with 3D QSVT, several researchers have successfully acquired more accurate information regarding viral processes in live cells.
Thus, the QSVT technique enables researchers to follow the fate of individual viruses interacting with different cellular structures from milliseconds to hours, providing more accurate and detailed information about viral infections in live cells. By complementing with cell labeling techniques and drug inhibition assays, this technique allows extensive access to dynamic information about the cellular components associated with viruses to reveal their underlying biological mechanisms.
You can read more about our approach and methods in the paper in Nature Protocols, https://www.nature.com/articles/s41596-022-00775-7.
Follow the Topic
-
Nature Protocols

This journal publishes secondary research articles and covers new techniques and technologies, as well as established methods, used in all fields of the biological, chemical and clinical sciences.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in