Social interactions in biofilms shaped by awoken mobile genetic elements
Published in Ecology & Evolution

An intriguing feature of bacterial biofilms is that the matrix encased cells are influenced by the complex interactions shaped by local chemical heterogeneity. In addition, this spatial scale greatly impacts the evolution of microbes. For a microbiologist, studying the evolution of biofilms offers a fascinating approach to understand the importance of its structural components. For an evolutionary biologist, the simplicity and rapid growth of microbial biofilms render a perfect system to test theoretical predictions on exploitability and evolutionary stability of matrix components.
Our research group is engaged with understanding the life of the Gram positive bacterium, Bacillus subtilis. This bacterium is prone to attach to various surfaces, including the plant rhizosphere, where it protects the plant from pathogenic microbes. Attachment and subsequent biofilm formation are triggered by plant secreted compounds or plant-derived polysaccharide molecules. Other type of biofilms, termed pellicles can be observed on the air-medium interface. B. subtilis pellicles can also stick to plastic surfaces. When a robust pellicle is developed in a petri dish, the wrinkles of the well-developed pellicle can occasionally reach the top of the plate and stick to it (see video). These pellicles depend on the production of an extracellular matrix, polymers that are secreted out from the cells and possibly shared among the cells of the pellicles. Therefore, the pellicles of B. subtilis represent an intriguing model to study social interactions based on the matrix production.

The initial goal of our project was to examine
the impact of non-matrix producing strains (NMPs) on the matrix producing wild
types (MPs) in a pellicle and to perform an experimental evolution on this
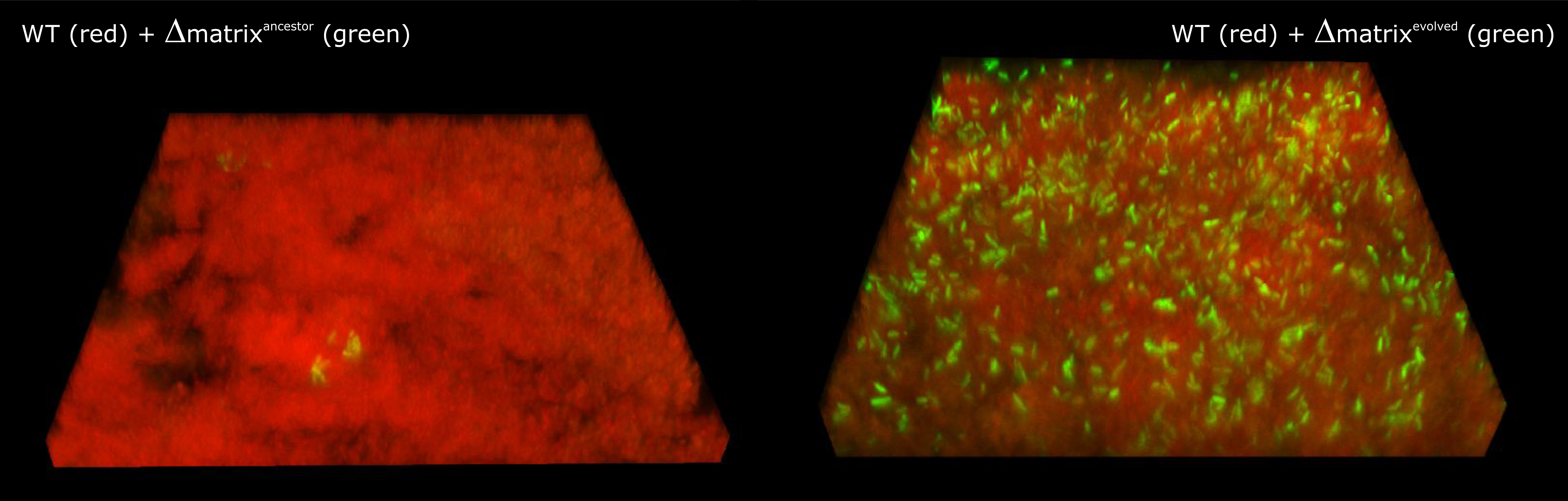
co-culture system. While this seemed a good idea, strikingly, these NMPs were
unable to incorporate into the pellicles made by MPs. Instead of concluding the
laboratory evolution approach and turning to a different methodology, we
continued to subsequently cultivate the mixed populations of MPs and the remaining
NMP minority.
To our surprise, after several transfers, the level of NMPs increased in several populations. This observation started a plethora of laborious experiments (including the use of thousands of plates for fitness assays) designed to scrutinize what had happened in these mixed cultures.

Since the increased NMP abundance was observed
in relatively short time (only 10 transfers), and we had access to great high
throughput sequencing techniques, we expected to rapidly identify the mutations
responsible for spread of NMPs (eNMPs) in the pellicles. Again, the genome
re-sequencing of evolved populations and isolates yielded a pretty surprising outcome
at the genetic level: about 1000 mutations were mapped on the genome, localized
mostly in prophage regions. Interestingly, the mutations were identified both
in eNMPs and eMPs and the target genes showed almost perfect overlap. While
this huge number of mutations was unpredicted, it created the opportunity for series
of innovative experiments that led us to the discovery how the de novo evolved phage weaponry
contributed to the shift in the social interaction between eNMPs and eMPs
within the pellicles.
Our experiments revealed an intriguing and
complex microbial system involving the social exploitation of shared biofilm
matrix and awakening of sleeping prophages. It will be interesting to see
whether these evolutionary innovations observed in simple single-species setup
can also be witnessed in ecological settings, e.g. when B. subtilis biofilms colonize the plant rhizosphere. We believe
that the ecology of phages might have more impact on the evolution of microbial
social interactions than previously anticipated.
Martin M*, Dragoš A*, Hölscher T*, Maróti G, Bálint B, Westermann M, Kovács ÁT (2017) De novo evolved interference competition promotes the spread of biofilm defectors. Nature Communications 8:15127. doi: 10.1038/NCOMMS15127 (*co-first authors), available here: http://go.nature.com/2prcjJu
(Cover image created by Eisha Mhatre)

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in