Synthetic biology - recombineering proteins to generate cellular powerhouses
Published in Bioengineering & Biotechnology, Protocols & Methods, and Cell & Molecular Biology

Synthetic biology is an avant-garde field of nascent multidisciplinary research that primarily aims to re-engineer cells as industrial machines. I have always found the concept and the process of rewiring gene networks of a natural biological cell for customized behavior to be promising (and simultaneously fascinating) in the fields of biomedicine and biotechnology. Previously, I made notes looking into some of the advances of gene circuit engineering, and of gene editing and recombineering methods that are built on the futuristic goals of rebuilding a cell to carryout human designed functions (Figure 1) [Healy 2019]. These efforts provide a window to interrogate the pathophysiology of cells, to then design and develop therapeutic interventions, to highlight the epitome of ambition in biology. Since I recently emphasized the significance of proteomics relative to detecting culprit proteins that underly the origin of disease, this post initially looks into recombineering proteins through synthetic biology, to briefly highlight mechanistic details of the biomolecular engine.
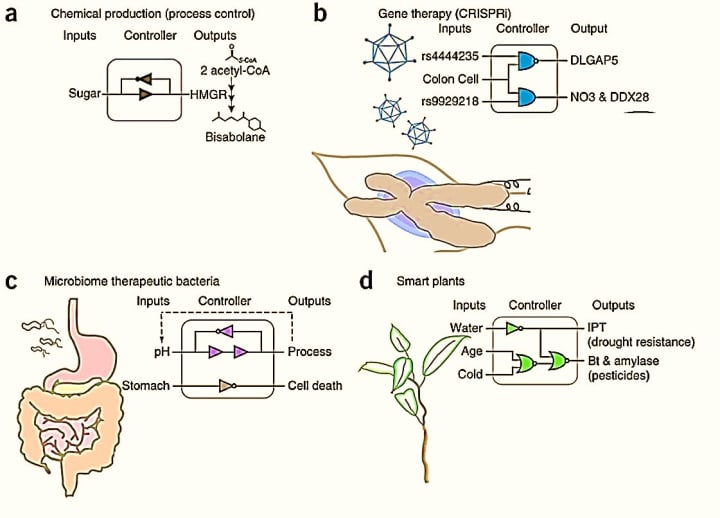
Figure 1: Hypothetical applications of gene circuit engineering: a) chemical production of non-toxic alternatives to diesel, b) gene therapy for early detection and to edit genetic anomalies in diseases, c) engineering therapeutic gut bacteria, d) smart plants that sense environmental cues to implement responses. Credit: [Brophy 2014].
Synthetic biology is a very young field relative to the advent of gene circuit engineering that debuted in the year 2000, with bottom-up engineering approaches that can assemble genetic parts into more complex gene circuits to facilitate new functionalities into cells (Figure 2) [Healy 2019]. Such advances include ‘smart plants’ and therapeutic agents that can correct genetic diseases, most of these ideas must expand beyond their hypothetical stage to meet the test of time [Brophy 2014]. Very briefly, gene circuit development depends on modularity, where the composing units do not change upon interconnection, allowing scientists to predict gene circuit behaviors. The process of upgrading protein synthesis for synthetic biology on the other hand, depends on the expansion of the genetic code [Donoghue 2013], and this therefore forms the primary focus here.
This post ventures to describe the development of non-standard and non-canonical amino acids that are either chemically modified after their incorporation into a protein via posttranslational modifications or are present as organic molecules containing an amino and a carboxylic acid, to form ‘peptidomimetic’ building blocks [Castro 2023]. Unnatural amino acids or non-canonical amino acids are not located naturally in the genetic code of organisms and are either found in nature as intermediates of biosynthesis pathways or are developed synthetically in the lab [Chen 2010]. Such designs can be used to form new bioactive molecules and new biomaterials. The post examines a few methods of gene editing and recombineering to rewire cells, to understand and intervene pathological cascades for diagnostics and therapeutic applications.
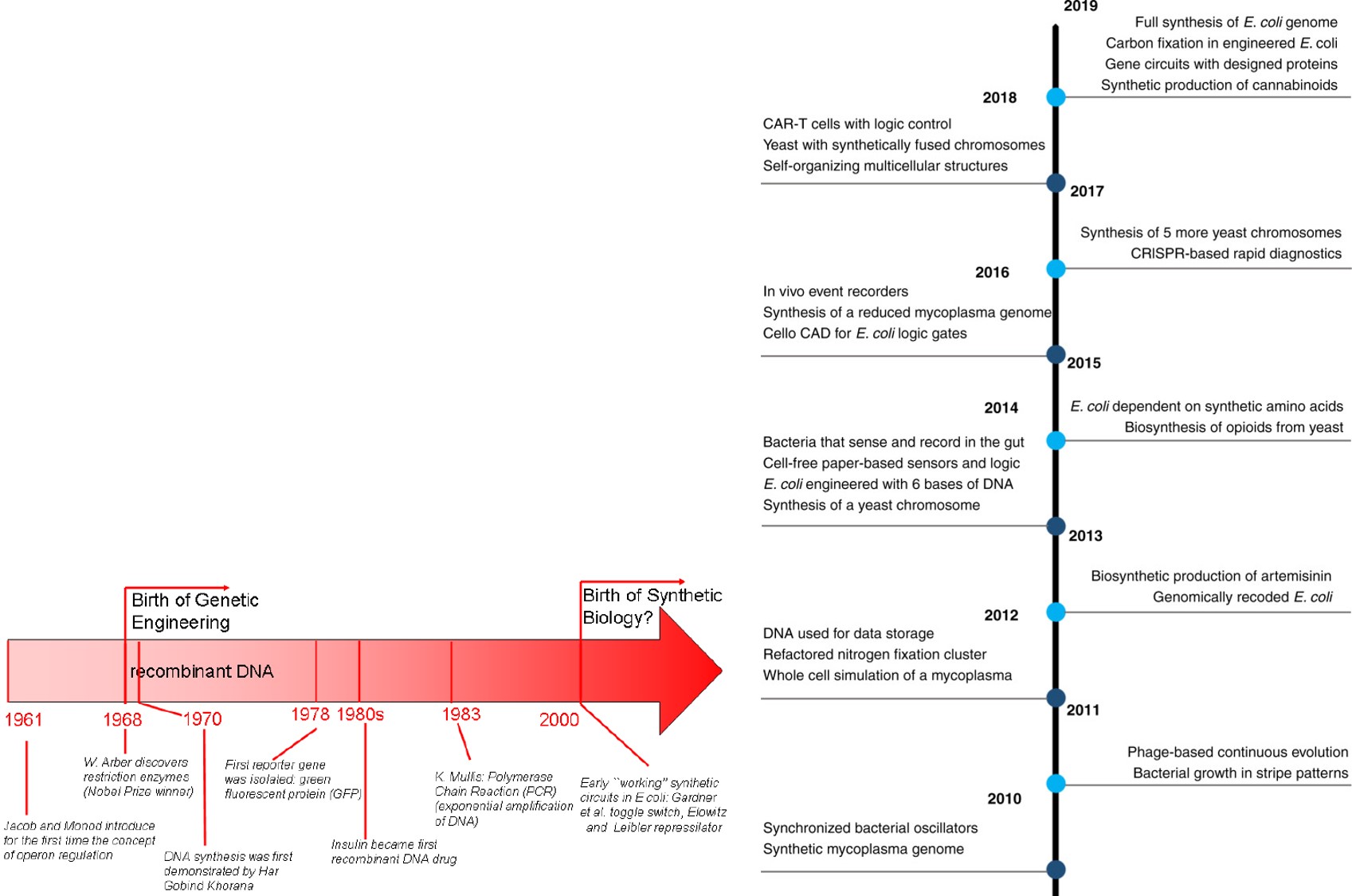
Figure 2: Milestones in the history of synthetic biology and landmark research achievements span from 1961 to 2020. The advent of synthetic biology relative to synthetic circuits is attributed to the year 2000. Each timeframe presents brief summaries of some key research milestones. Credit: [Del Vecchio 2009, Meng 2020].
The origin of the genetic code and its ‘non-frozen’ nature.
The origin of the genetic code takes us to the early years, when Francis Crick, James Watson and Rosalind Franklin were famously investigating the blueprint of life and the architecture of the DNA. In a Letter to Nature, Crick postulated the genetic code to have originated from a ‘frozen accident’ [Crick 1967, Ambrogelly 2007]. This concept thawed with time, to reveal the non-immutable or non-frozen nature of the genetic code that came to light with several findings, for instance in the 1970s, the stop codon was reassigned to encode tryptophan in yeast and in mitochondria [Macino 1979]. In nature, all 64 codons in the genetic code can be assigned to 20 different kinds of proteinogenic amino acids and three stop codon signals (ochre UAA, umber UGA, and amber UAG) during protein translation (Figure 3) [Chavatte 2003]. Similarly, it is possible to incorporate non-standard amino acids (NSAAs) in the lab, for enhanced properties of new proteomics for diverse applications [Lajoie 2013].
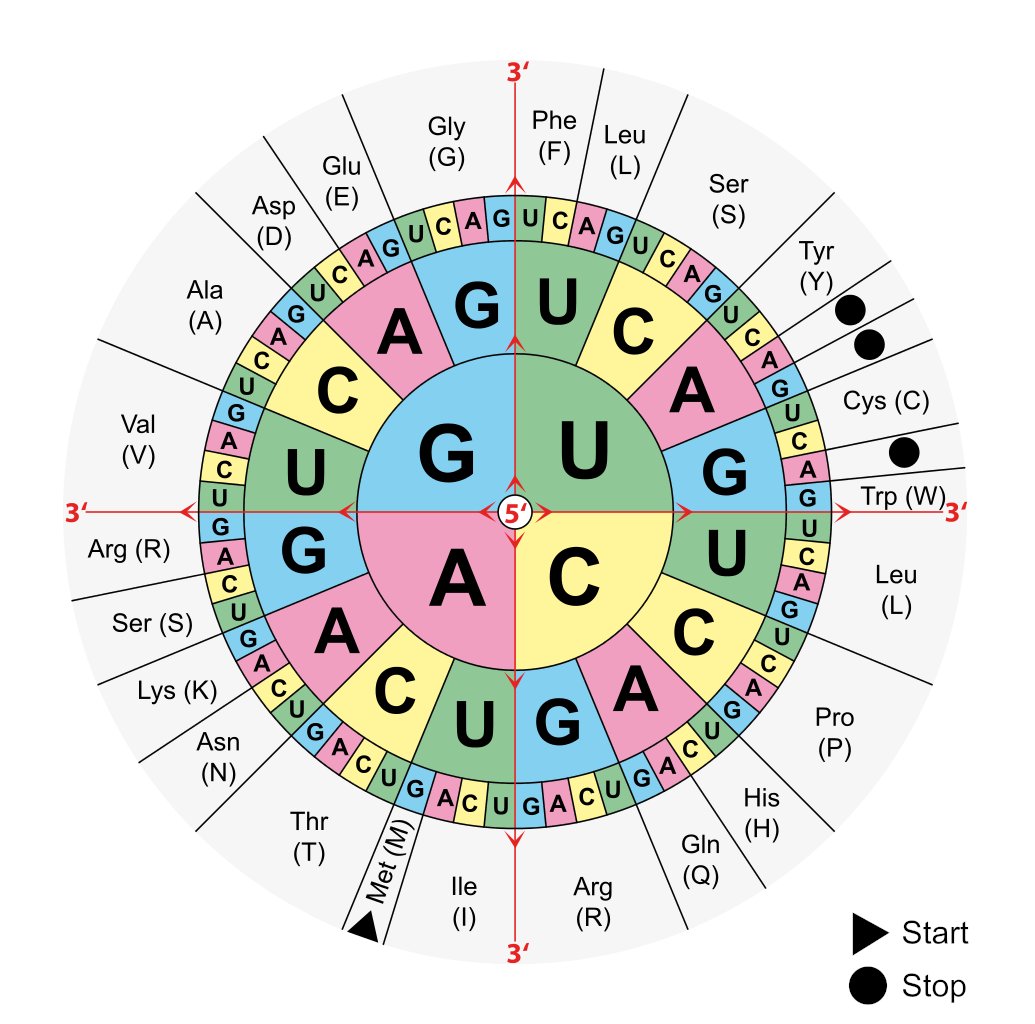
Figure 3: The wheel of codons used to translate a genetic code into a sequence of amino acids. The 64 codons encode 20 amino acids. Credit: [Ambrogelly 2007].
The next big surprise in proteomics came to be in the 1980s, when selenocysteine was discovered as the 21st genetically encoded amino acid by recoding the UGA stop codon for its incorporation in many species, including humans [Böck 1988]. The synthesis of selenocysteine is specifically targeted for human health and development [Bell, 2024]. The 22nd amino acid encoding pyrrolysine was discovered in 2002 [Hao 2002, Gaston 2011] thus overcoming the assumption of the immutability of the genetic code. While the genetic code can evolve, its translation can be rewired to genetically encode more than the 20 standard amino acids by recoding the stop codon.
Protein synthesis with synthetic biology – adding new chemistries to the genetic code
Expanding the genetic code is a definitive goal in synthetic biology with an established amount approximating 100 distinct noncanonical amino acids (ncAAs) generated via orthogonal translational systems. The process can allow the facile in vitro production of proteins with hardwired post-translational modifications to include unnatural amino acids containing a photo-cage or photolabile protecting group, or site-specific fluorescent labels for further expansion [Liu 2010]. There are limits to the natural protein synthesis machinery that must be understood at a mechanobiology level to further rewire the genetic code for expansion in this way. Protein synthesis for synthetic biology can expand the genetic code beyond the 20 canonical amino acids via specific-orthogonal translational systems that must optimize the following factors (Figure 4):
- An ‘open’ codon to encode
- Noncanonical amino acids that can permeate the cell.
- An aminoacyl-tRNA synthetase to efficiently ligate a desired non-canonical amino acid,
- A tRNA that can decode the ‘open’ codon, and
- Compatible elongation factors and ribosomes
Tailor-made genetic codes can be synthetically produced by using a reconstituted in vitro translation system to facilitate the synthesis of unnatural amino peptides with unmatched flexibility [Jewett 2016]. Geneticists can accomplish this by preparing the genetic code by engineering an efficient orthogonal translational system with the illustrated components (Figure 4).
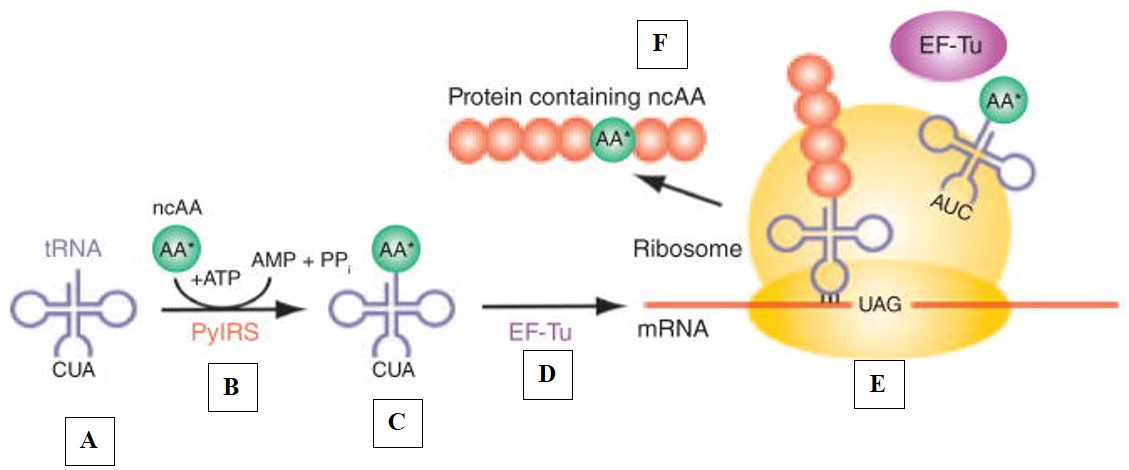
Figure 4: The mechanism-of-action of protein synthesis requires amino acid-tRNAs as building blocks for ribosomal protein synthesis. Each stage of synthesis, is labelled from A-F, (A) Amino acid-tRNAs are in use to form building blocks for ribosomal protein synthesis, (B) The amino-acid can be ligated to the tRNA by using a dedicated enzyme to develop a protein, subsequently this leads to (C) The assembly of a protein complex, which leads to (D) The development of a product of amino acid-tRNAs delivered by an elongation factor such as EF-TU to the ribosome. (E) Once inside the ribosome, the anticodon of the tRNA matches the triplet codon of the mRNA, this then leads to (F) Protein synthesis to create an expanded genetic code that incorporates the unnatural peptides. Credit: [Donoghue 2013].
When compared to the use of natural peptide drugs, the strategies described here have successfully established site-directed insertion of noncanonical amino acids into proteins to generate ‘peptidomimetics’ and overcome the problems of pharmacokinetics and enzymatic stability [Donoghue 2013, Castro 2023] (Table 1). To develop more optimal orthogonal pairs, novel genomic recombineering methods such as multiplex automated genome engineering (MAGE) and clustered regularly interspaced short palindromic repeats (CRISPR) offer better options [Wang 2009, Doudna, Charpentier 2014]. The high-throughput capacity for structure-guided design and rational mutagenesis plays a significant role to engineer enzymatic activity and successfully redesign active sites for broad-ranging biomedical applications.
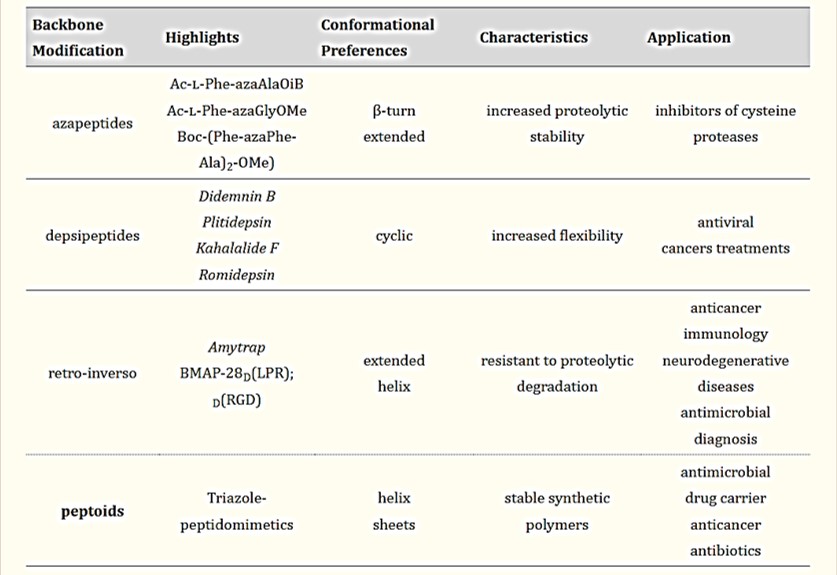
Table 1: Backbone modifications – biological applications and preferable secondary structure of peptidomimetics for precision medicine [Castro 2023].
Accelerated evolution - versatile methods of genomic diversity
One of the challenges of bioengineering is to disentangle specific factors that encode observed biological behaviors. Scientists often accomplish this by perturbing the cell to measure the effects of agitations on cellular physiology. Thus far, the development of new orthogonal aminoacyl-tRNA synthetase/tRNA pairs has led to the addition of approximately 70 unnatural amino acids to the genetic codes, in model organisms such as Escherichia coli, yeast, and mammalian cells (Figure 5). Escherichia coli is the workhorse organism of most synthetic biology experiments and is a widely used expression host [Liu 2010].
In fact, I briefly detailed bacterial expression systems within a practical setting with Escherichia coli to recombineer the human protein tropoelastin, when describing the ‘then nascent era’ of synthetic biology, on a previous post published here on Nature Portfolio. The increasingly multidisciplinary nature of modern science can promote the innovation of new theories and techniques with synthetic biology spearheading the niche [Meng 2020, Zhao 2023]. MAGE or multiplex automated genome engineering, for instance, represents a set of highly multiplexed single-stranded DNA-mediated methods of recombineering that were also first described in E. coli, and rapidly transferred into diverse prokaryotes and eukaryotic cell lines [Wang 2009, Wannier 2021].
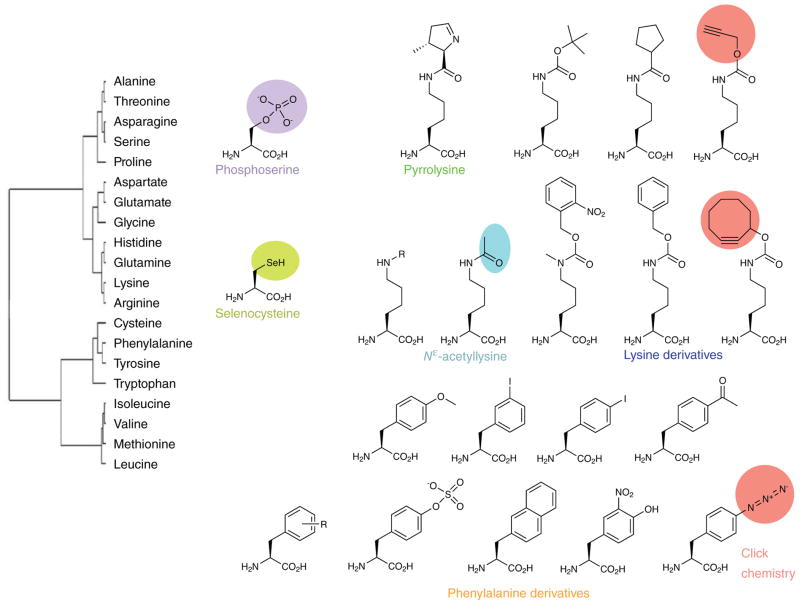
Figure 5: Chemical diversity of amino acids in the standard and expanded genetic codes. Amino acid similarity reflects the substitution frequency of one amino acid for another in standardized sets of multiple sequence alignments. Credit: [Donoghue 2013].
Alongside synthetic biology, the sequential advent of systems biology can detail the high throughput -omics methods from genomics to transcriptomics, proteomics, and metabolomics for bottom-up engineering (another recently covered topic), to provide massive datasets that garner a more comprehensive understanding of complex biological networks, to facilitate the identification of pathological irregularities [Del Vecchio 2009, Donoghue 2013].
The advent of multiplex automated genome engineering (MAGE)
Most interdisciplinary concepts of genome engineering tease apart the workings of cellular processes to shed light on key biomolecular events governing the behavior of pathological cascades. For instance, MAGE in its mechanism-of-action can rapidly and continuously generate a diverse set of rapid genetic changes in a cyclic and scalable manner within cells (Figure 6) [Wang 2009]. By using the high-throughput technology, bioengineers have optimized the 1-deoxy-D-xylulose-5-phosphate biosynthesis pathway in E. coli, to overproduce an industrially significant isoprenoid lycopene as proof-of-concept [Lichtenthaler 1999].
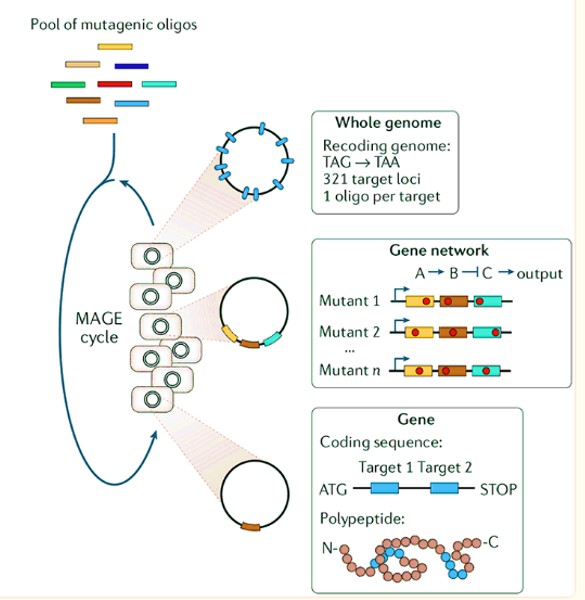
Figure 6: Multiplex automated genome engineering or MAGE enables the rapid and continuous generation of sequence diversity at many targeted chromosomal locations across a large population of cells through the repeated introduction of synthetic DNA. Each cell contains a different set of mutations, producing a heterogeneous population of rich diversity. Credit: [Wang 2009, Wannier 2021]
The isoprenoid superfamily of compounds are precursors of metabolic pathways; integral in commercial biotechnology to deliver therapeutics, nutraceuticals, and fine chemicals [Klein-Marcuschamer 2007]. The premise of engineering ‘microbial cell factories’ has a stronghold in metabolic engineering, with broad-ranging impact across basic research and industry applications. A classic example of this complexity details the synthesis of isoprenoids beyond lycopenes by combining synthetic biology and systems biology for metabolic engineering in microbial cells [Klein-Marcuschamer 2007]. These early efforts have collectively seen through to bioengineer new compounds via protein engineering or genetic engineering methods [Tobias 2006].
Mechanisms-of-action: genome editing without inducing double-strand breaks
The MAGE method relies on oligonucleotide-mediated allelic replacement at multiple sites, and the introduction of synthetic DNA to facilitate genetic modifications at high efficiency within favorable evolutionary paths [Carr 2012]. Several years after the MAGE proof-of-concept study was conducted in E. coli, a research team incorporated the same method with eukaryotes to precisely edit multiple sites in Saccharomyces cerevisiae, without generating DNA double-strand breaks [Wrighton 2017, Barbieri 2017]. This is a significant feature, since the process of multisite editing is typically limited by double-strand break mechanisms due to three reasons (Figure 7):
- First, cleaving the genome is cytotoxic, and cell lethality increases when double-strand breaks are introduced across multiple target sites.
- Second, in eukaryotes double strand break repair is subject to additional, unwanted insertions or deletions for cleavage even after editing,
- Third, it is difficult to simultaneously modify many loci in a single cell due to the inefficiency of generating targeted single base pair edits with double-strand breaks.
Creating precise edits at single base pair resolution in eukaryotes is thus challenging via DNA double-strand breaks for applications of gene correction and genetic disease treatment [Barbieri 2017]. Most genetic diseases arise from point mutations. Methods that are in use to correct such defects nevertheless continue to rely on inefficient double strand DNA breaks. Generally, CRISPR-Cas9 induced double-strand breaks and non-homologous end joining repair offer a knock-in strategy for the site-specific introduction of nonnatural chemical groups into proteins in living cells [Meineke 2023]. While Cas9-guided deamination can edit DNA base-pairs efficiently without inducing a double strand break, the method is limited to specific mutations alone [Komor 2016].
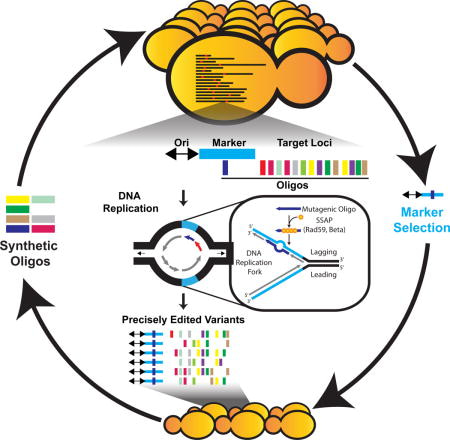
Figure 7: Replication forks can be co-opted to introduce multisite mutations in eukaryotes, without the need for double-strand breaks [Barbieri 2017].
The multisite genetic editing technique of MAGE for eukaryotic genome engineering thus varies from most other eukaryotic genome editing methods such as Zinc finger nucleases and CRISPR-Cas9 [Doudna, Charpentier 2014], to introduce genomic modifications without creating double-strand breaks. While earlier methods of zinc finger nucleases (ZFNs) and transcription activator-like efficient nucleases (TALENs) recognize DNA sequences of interest and identify protein-DNA interactions to induce the usual double-strand breaks at genomic loci, their construction too remains laborious and costly [Barbieri 2017].
Comparatively, MAGE can be automated to generate large numbers of precise edits into a single cell during many cycles, for molecular evolution of single genes within cells for a lofty goal of re-engineering cells as powerhouses for synthetic biology. While this technique can recode the whole genome in eukaryotes, it can target viral genomes, bacteria, plasmids, and artificial chromosomes too [Gallagher 2014]. The goal is to tinker MAGE to overcome its inherent limitations and arrive at an optimal version for multisite genome editing of stem cells to treat cancer and other rare diseases, while expanding its industrial portfolio [Bohannon 2011, Singh 2015].
Bioengineering complex biological systems – from the lab to the clinic
Synthetic biology is accurately defined as a field of risk-takers, where researchers from multidisciplinary areas come together to design and develop ‘wild experiments,’ to hack complex biological systems and arrive at solutions to advance the fields of medicine and industrial biotechnology [Bohannon 2011]. This view is exemplified on this post, by highlighting multiple site editing techniques to change the function of a cell or of a protein, by simply manipulating its genetic code.
Decades of synthetic biology have seen to the development of the most hyped topics of the century, with landmark achievements that highlight the pinnacle of human invention [Meng 2020]. The preceding decade has seen standout technologies including Cello; an end-to-end computer-aided design platform for logic circuit construction in E. coli designed by Christopher Voigt and team [Neilsen 2016], as well as in vivo event recorders using DNA as a memory device to track biological events across time [Sheth 2018], and the capacity to use DNA for data storage, beyond genomics for archival potential [Church 2012].
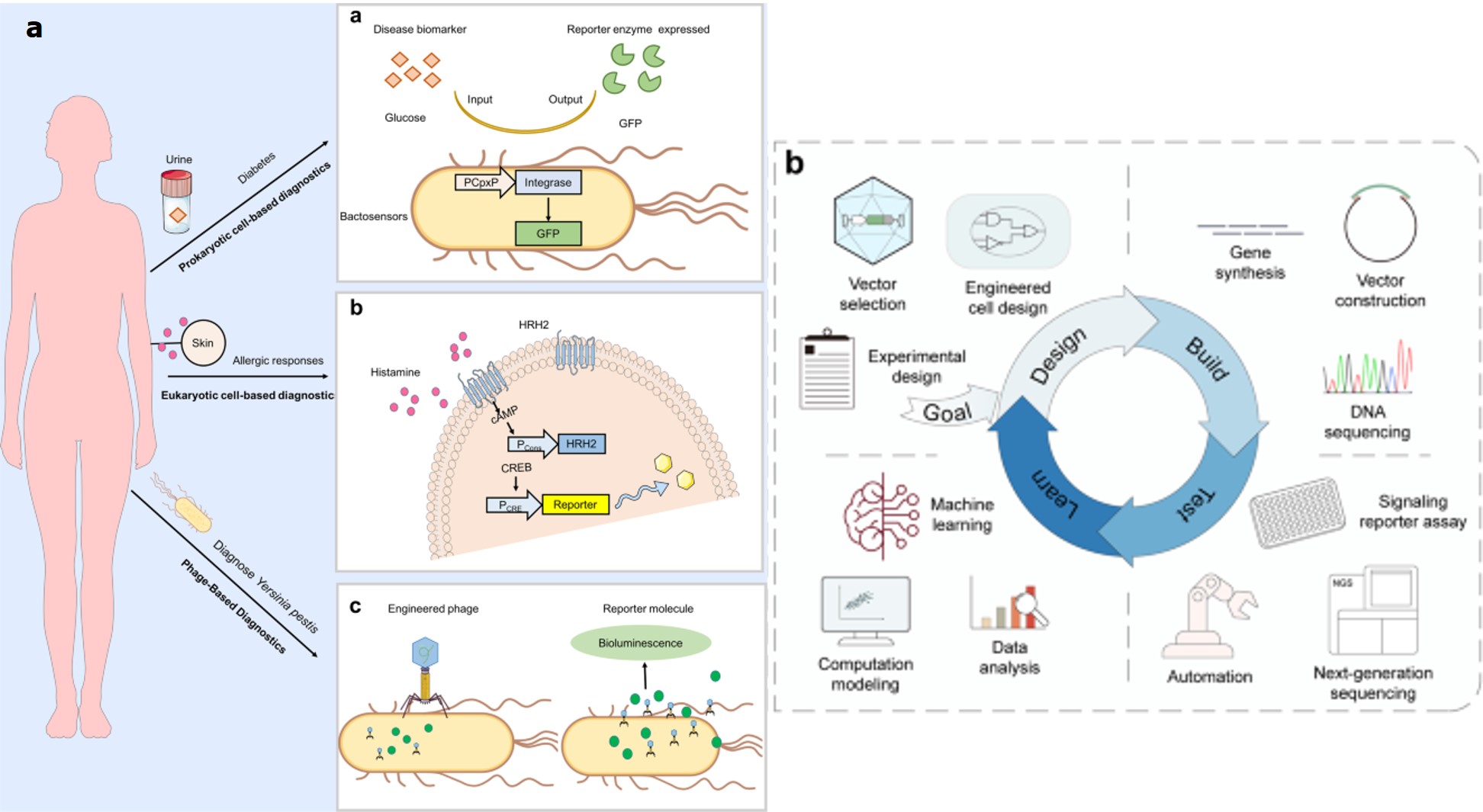
Figure 8: A) synthetic biology platforms for diagnostics: platforms a-c indicate applications of engineered bacteria, mammalian synthetic biology, and phage-based diagnostics in sensing disease and producing a diagnostic readout. B) A modified design-build-test-learn-based framework can direct researchers to choose or engineer synthetic receptors for their applications in cell therapy or gene therapy. The ‘goal’ defines the design objectives for engineered cell or gene therapy, for clinical applications. Credit: [Zhao 2023, Teng 2024].
Among the variety of existing possibilities, I have focused on the integration of biotic signals such as ions, metabolites, nucleic acids, or proteins involved in signaling cascades, for genetic and epigenetic regulation at transcriptional, translational, and post-translational stages, to develop robust cell behaviors. The most promising practical aspects of the existing hype apply to cell and gene therapies, to treat and correct a range of rare diseases [Teng 2024]. The niche is rampant with targeted and personalized treatments such as CAR T (chimeric antigen receptor T) cell therapies to treat blood cancers [Kimbrel 2020], hematopoietic stem cells engineered to correct hematological disorders [Ferrari 2021], and gene therapies to treat spinal muscular atrophy and restore vision - that are already approved for clinical use [Cehajic-Kapetanovic 2023].
More recent efforts in the field of biomedicine have seen delicately designed and engineered synthetic receptors to regulate the function of therapeutic cells to fine-tune therapeutic intervention, alongside user-defined signals or biomarkers for clinical translation (Figure 8) [Teng 2024]. Synthetic receptors notably include CARs (chimeric antigen receptors) with a long-standing history of landmark achievements generated with natural or artificial components to endow designer cells with custom functions, to rewire the cellular input-output relationships [Teng 2024]. Such designer cells can be of mammalian origin to sense and respond to a variety of disease biomarkers to trigger downstream signaling and fine-tune custom therapeutic effects. By humanizing synthetic receptors, it is possible to expand their therapeutic applications through stem-cell engineering for cancer immunotherapy, to develop next-generation medicine [Li 2021].
The work here describes a very small fragment of the vast potential and promise of a growing multidisciplinary field with ample scope to understand the impact of synthetic biology in medicine and biotechnology. The research field is a fertile ground to devise and develop unprecedented experimental ideas for rapid progress in gene therapy and commercial biotechnology that have hitherto remained unreachable by established approaches.
Header Image: Gene editing and ethics, via the Harvard Magazine.
References
- Healy C. P. et al. Genetic circuits to engineer tissues with alternative functions, Journal of Biological Engineering, 2019.
- Brophy J. et al. Principles of gene circuit design, Nature Methods, 2014.
- O’Donoghue P. et al. Upgrading protein synthesis for synthetic biology, Nature Chemical Biology, 2013.
- Castro T. et al. Non-canonical amino acids as building blocks for peptidomimetics: structure, function, and applications, Biomolecules, 2023.
- Chen I. et al. Quadruplet codons: One small step for a ribosome, one giant leap for proteins, Bioessays, 2011.
- Crick J. et al. An Error in Model Building, Nature, 1967
- Ambrogelly A. et al. Natural expansion of the genetic code, Nature Chemical Biology, 2007.
- Macino G. et al. Use of the UGA terminator as a tryptophan codon in yeast mitochondria, PNAS 1979.
- Chavatte L. et al. Stop codon selection in eukaryotic translation termination: comparison of the discriminating potential between human and ciliate eRF1s, the EMBO journal, 2003.
- Lajoie M. et al. Genomically recoded organisms expand biological functions, Science, 2013.
- Böck A. et al. Selenocysteine, a highly specific component of certain enzymes, is incorporated by a UGA-directed co-translational mechanism, Biofactors, 1988
- Bell H. et al. Ironing out the role of ferroptosis in immunity, Immunity, 2024.
- Hao B. et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase, Science, 2002.
- Gaston M. et al. Functional context, biosynthesis, and genetic encoding of pyrrolysine, Current Opinions in Microbiology, 2011.
- Liu C. et al. Adding new chemistries to the genetic code, Annual Review of Biochemistry, 2010.
- Jewett M. et al. Tailor-made genetic codes, Nature Chemistry, 2011.
- Wang H. et al. Programming cells by multiplex genome engineering and accelerated evolution, Nature, 2009.
- Doudna J. and Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9, Science, 2014.
- Meng F. et al. The second decade of synthetic biology: 2010–2020, Nature Communications, 2020.
- Zhao N. et al. Synthetic biology-inspired cell engineering in diagnosis, treatment, and drug development, Signal Transduction and Targeted Therapy, 2023.
- Wannier T. et al. Recombineering and MAGE, Nature Review Methods Primers, 2021.
- Del Vecchio D. et al. Synthetic Biology: A Systems Engineering Perspective, MIT Press Direct, 2009.
- Lichtenthaler H. et al. The 1-Deoxy-D-Xylulose-5-Phosphate pathway of isoprenoid biosynthesis in plants, Annual Review of Plant Physiology and Plant Molecular Biology, 1999.
- Marcuschamer K. et al. Engineering microbial cell factories for biosynthesis of isoprenoid molecules: beyond lycopene, Trends in Biotechnology, 2007.
- Tobias A. V. et al. Biosynthesis of novel carotenoid families based on unnatural carbon backbones: A model for diversification of natural product pathways, Biochimica et Biophysica Acta, 2006.
- Carr P. et al. Enhanced multiplex genome engineering through co-operative oligonucleotide co-selection, Nucleic Acid Research, 2012.
- Wrighton K. et al. Multiplex genome engineering in eukaryotes, Nature Reviews Genetics, 2017.
- Barbieri E. et al. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes, Cell, 2017.
- Meineke B. et al. Generation of amber suppression cell lines using CRISPR-Cas9, Methods in Molecular Biology, 2023.
- Komor A. et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage, Nature, 2016.
- Gallagher R. et al. Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA, Nature Protocols, 2014.
- Bohannon J. et al. The Life Hacker, Science, 2011
- Neilsen A. et al. Genetic circuit design automation, Science, 2016.
- Sheth R. et al. DNA-based memory devices for recording cellular events, Nature Review Genetics, 2018.
- Church G. et al. Next-Generation Digital Information Storage in DNA, Science, 2012.
- Teng F. et al. Programmable synthetic receptors: the next-generation of cell and gene therapies, Nature, 2024.
- Kimbrel E. et al. Next-generation stem cells - ushering in a new era of cell-based therapies, Nature Reviews Drug Discoveries, 2020.
- Ferrari G. et al. Gene therapy using haematopoietic stem and progenitor cells, Nature Reviews Genetics, 2021.
- Cehajic-Kapetanovic J. et al. Bioengineering strategies for restoring vision, Nature Biomedical Engineering, 2023.
- Li Y. et al. Engineering stem cells for cancer immunotherapy, Cell Press, 2021.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in