Tasty, Therapeutic or Toxic? Silicon Nitride Nanopores as a Single Molecule Glycomics Sensor
Published in Chemistry
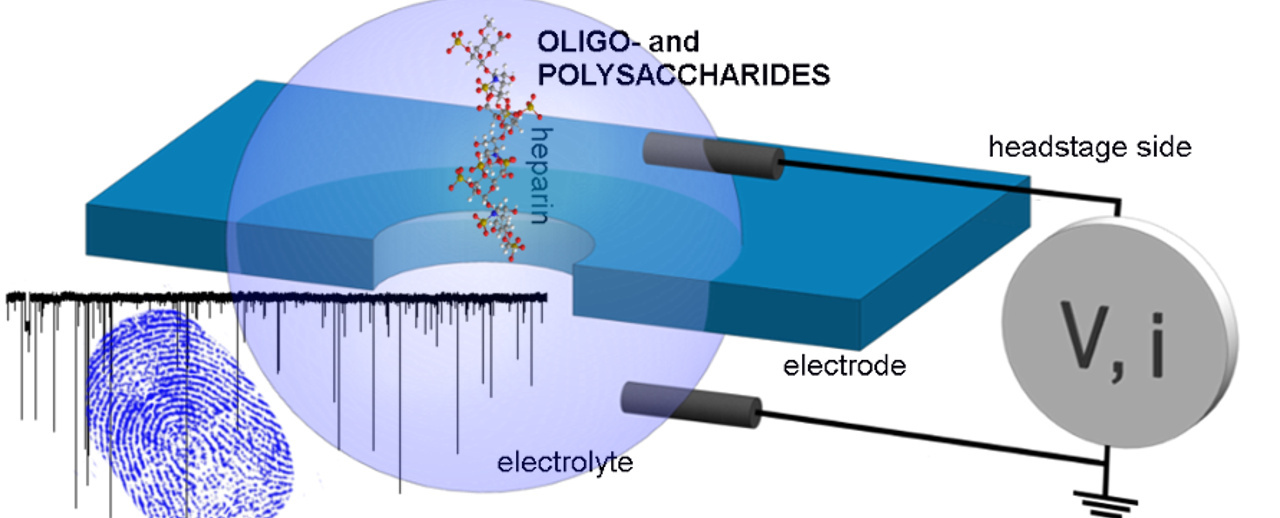
Our research group is focused on developing platforms for single molecule sensing, especially nanopores. A nanopore is a nanometer-diameter channel (on par with the size of single biomolecules) which can function as a miniature Coulter counter, only for molecules instead of particles. Since the initial discovery of nanopores as sensors in 19961, nanopores have shown immense potential for DNA and protein profiling, with the tremendously challenging study of complex polysaccharides in its early days. While many advanced and reliable characterization technologies have seen the light of day for DNA and proteins, e.g.: nuclear magnetic resonance, mass spectrometry, crystallography, polysaccharide profiling is still in its infancy.2 Compared to 4 nucleotide bases that make up DNA or 20 amino acids that make up proteins, there are ~120 naturally occurring monosaccharides that the glycomics researcher must contend with. Adding to the complexity are the different monomer linkages, isomeric forms and varying degrees of branching. Glycomics has been described as being in dire need of new tools and techniques—to complement classical techniques—to provide additional information on polysaccharides.
Could nanopores provide the enabling capabilities required by glycomics for the much-awaited major leap to match the pace of genomics and proteomics?
If DNA and proteins can be profiled by nanopores, why not polysaccharides?
With the fixed size of biological nanopores, profiling branched polysaccharides is not feasible. With their size tunability and robust nature, solid-state nanopores—made from materials like silicon nitride (SiNx)—are the ideal candidate for the profiling of these complex, biologically important molecules. Our interest in developing solid-state nanopores for glycomics dates to 2010, and we spent several years establishing a foundation before starting work focused on the analytes, themselves, in 2015. Here we go!
Exploring and establishing experimental conditions for an entirely new class of molecules demanded considerable ground-up experimentation. The lack of amplification technique analogues to PCR for DNA, added to the challenge. Go ahead, read our paper to find out how we overcome these challenges! We were motivated by earlier work using different pores and different molecules to use controlled dielectric breakdown pores with a range of polysaccharides with quite different properties.3-10 We first studied a polysaccharide known as sodium alginate which has really interesting material science applications but that also holds delight for the fans of molecular gastronomy. When we started our studies, we noticed that polysaccharides don’t always play by the rules: this molecule travelled the wrong way! (at least in comparison to the nanopore DNA sensing world) This lead us to explore the effect of electrophoresis, electroosmosis and nanopore surface charge.
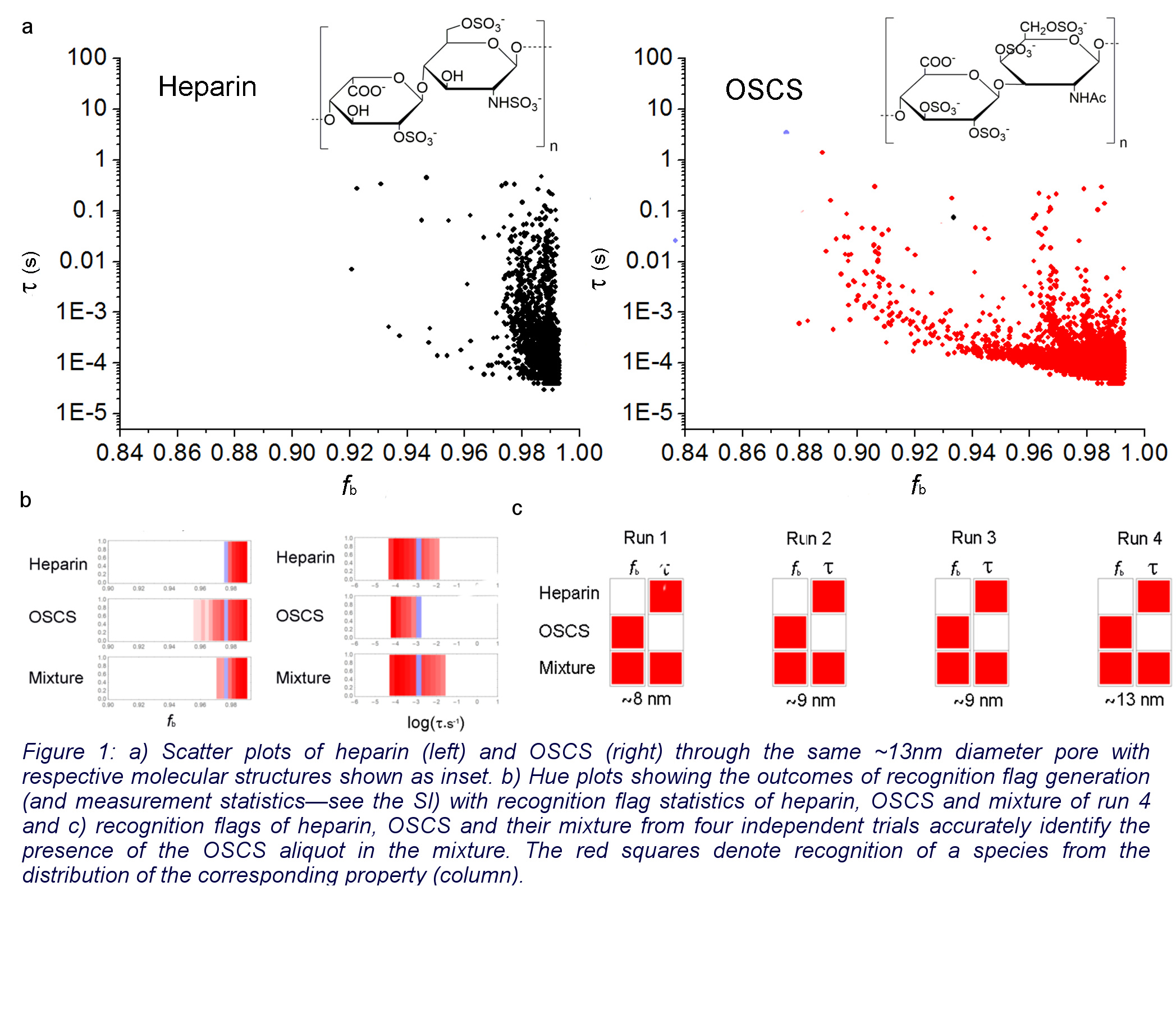
But we were fundamentally interested in studying heparin. Heparin is a globally used lifesaving polysaccharide-based anticoagulant drug. In 2008, there were severe heparin-related adverse effects due to the drug being contaminated by oversulfated chondroitin sulfate (OSCS)—a sugar structurally close to heparin—and failure to detect it resulted in over 100 deaths in the US. Our goal for this project was to ultimately explore the possibility of using a SiNx nanopore to detect the contaminant in heparin. After lot of fine tuning, two molecules revealed apparent differences in the event fingerprints despite the similar sizes and close resemblance in structure! After many trials with a range of nanopore sizes, we developed a program analysis method to identify and indicate OSCS being present in a heparin sample by analyzing the nanopore fingerprint and it proved successful over all our trials.
Heparin is not the first nor will it be the last incident of drug adulteration (or food adulteration for that matter) and nanopores could be saving our lives in the future! This is our first step towards uncovering the potential of solid-state nanopores as a tool for glycomics and we have much more exciting work yet to be done!
Find our Nature Communications paper Surveying silicon nitride nanopores for glycomics and heparin quality assurance here.
References
- Kasianowicz, J. J., Brandin, E., Branton, D. & Deamer, D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. 93, 13770–13773 (1996).
- National Research Council. Transforming Glycoscience: A Roadmap for the Future. The National Academies Press (2012).
- Bacri L, Oukhaled A, Hémon E, Bassafoula FB, Auvray L, Daniel R. Discrimination of neutral oligosaccharides through a nanopore. Biochem Biophys Res Commun 412, 561-564 (2011).
- Fennouri A, Przybylski C, Pastoriza-Gallego M, Bacri L, Auvray L, Daniel R. Single Molecule Detection of Glycosaminoglycan Hyaluronic Acid Oligosaccharides and Depolymerization Enzyme Activity Using a Protein Nanopore. ACS Nano 6, 9672-9678 (2012).
- Fennouri A, Daniel R, Pastoriza-Gallego M, Auvray L, Pelta J, Bacri L. Kinetics of Enzymatic Degradation of High Molecular Weight Polysaccharides through a Nanopore: Experiments and Data-Modeling. Anal Chem 85, 8488-8492 (2013).
- Kullman L, Winterhalter M, Bezrukov SM. Transport of Maltodextrins through Maltoporin: A Single-Channel Study. Biophys J 82, 803-812 (2002).
- Zhao S, et al. Sugar-stimulated robust nanodevice: 4-Carboxyphenylboronic acid modified single glass conical nanopores. Electrochem Commun 36, 71-74 (2013)
- Haywood, D.G., Saha-Shah, A., Baker, L.A. & Jacobson, S.C. Fundamental Studies of Nanofluidics: Nanopores, Nanochannels, and Nanopipets. Anal. Chem. 87, 172-187 (2015).
- Edel, J., Ivanov, A. & Kim, M. Nanofluidics. (The Royal Society of Chemistry, 2017).
- Bandara, Y.M.N.D.Y., Karawdeniya, B.I. & Dwyer, J.R. Real-Time Profiling of Solid-State Nanopores During Solution-Phase Nanofabrication. ACS Appl. Mater. Interfaces 8, 30583-30589 (2016).

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in