The antileukemic effect of methylated indolequinone MAC681 involves degradation of PARP1 and immunogenic necroptosis
Published in Cancer and Cell & Molecular Biology
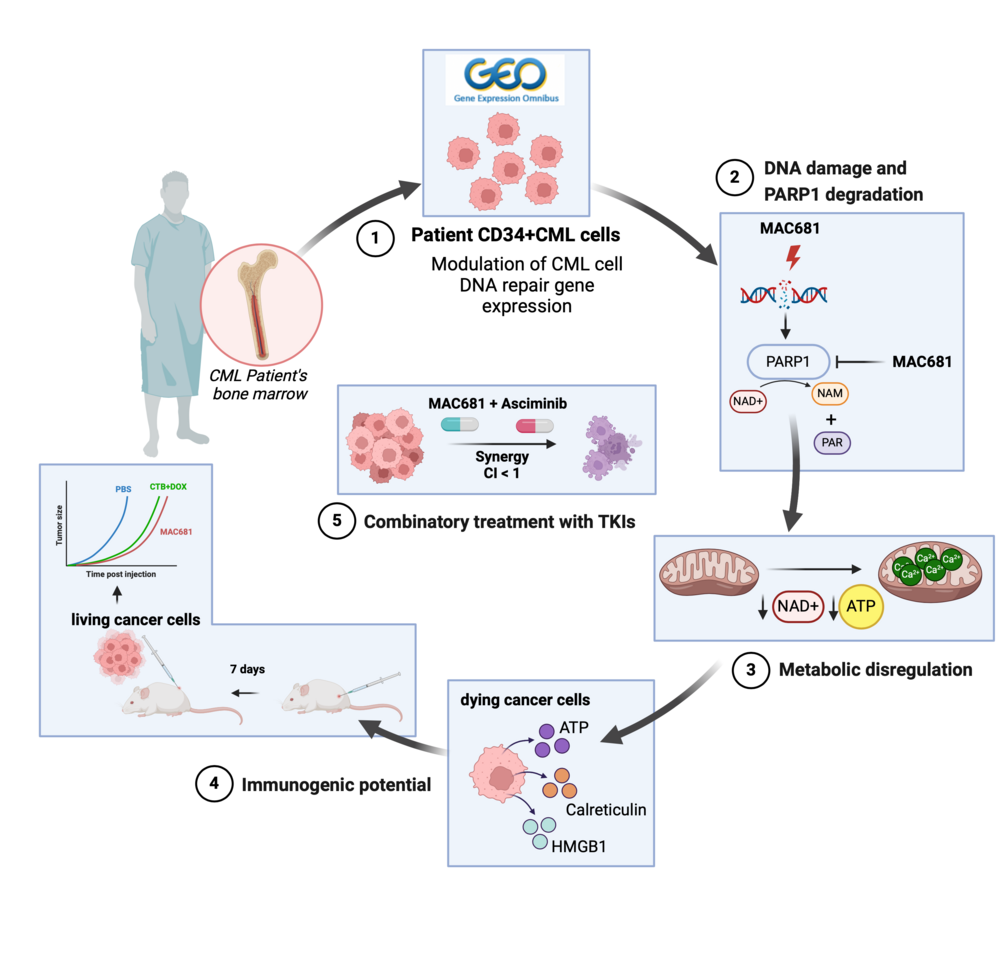
Chronic myeloid leukemia (CML), a blood cancer characterized by the BCR-ABL fusion gene, presents ongoing challenges despite advancements in treatment. While tyrosine kinase inhibitors (TKIs) target the constitutively activated kinase, drug resistance and the persistence of leukemia stem cells (LSCs) remain significant obstacles. Approximately one in five patients develop resistance, with 22% becoming intolerant and requiring treatment discontinuation due to side effects (1, 2). Persistent LSCs exhibit altered mitochondrial metabolism, rendering them resistant to conventional therapies (3). In response to these challenges, we investigated a novel approach targeting DNA damage and metabolic rewiring. We demonstrated the role of non-apoptotic cell death in the context of improved immunogenic therapies against CML. Our research shows the importance of damage-associated molecular patterns (DAMPs) release and their implications for the immune response in CML treatment.
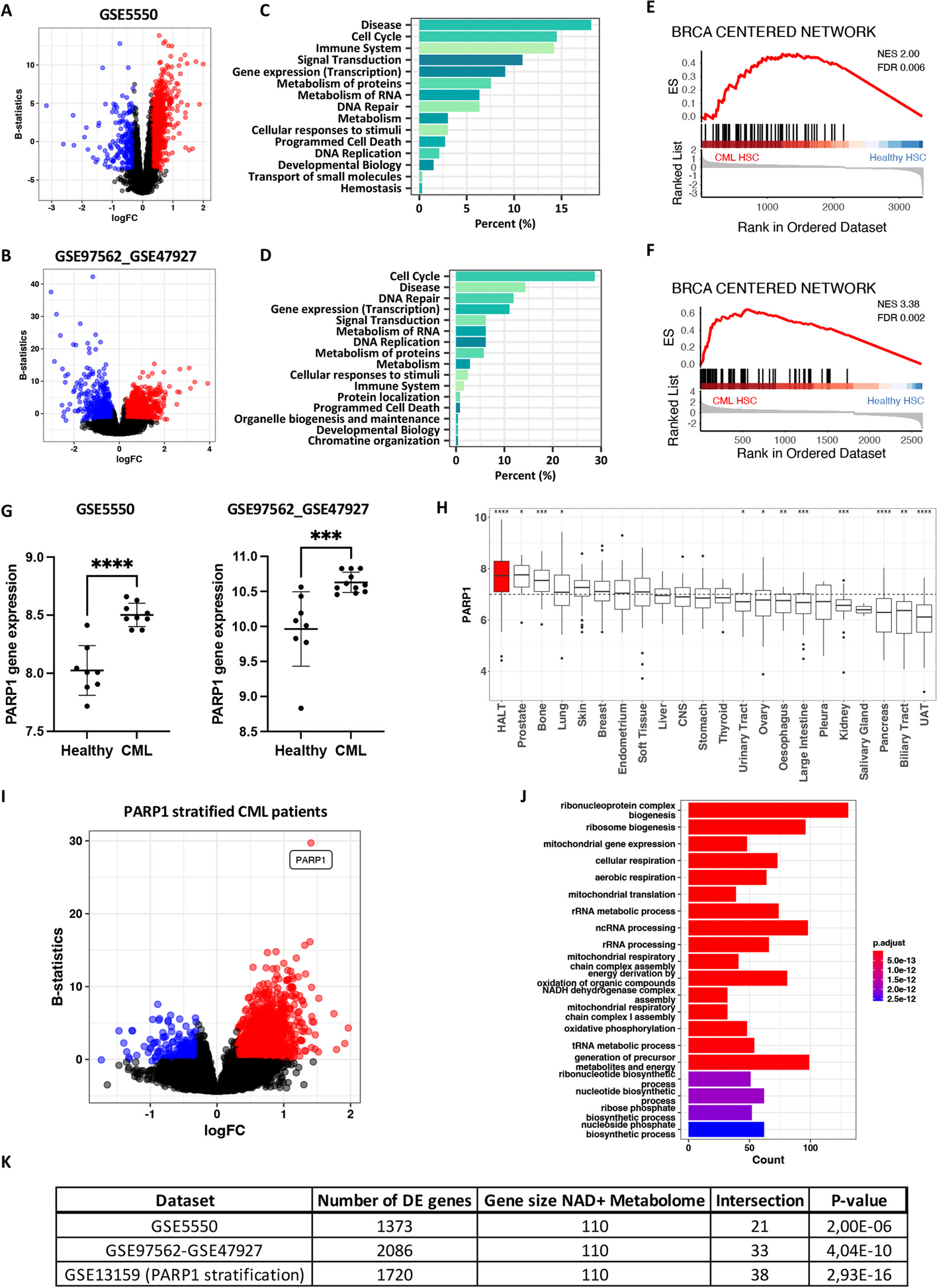
Our study began with an analysis of gene expression patterns in CML patients, revealing metabolic alterations and deficiencies in DNA repair pathways, including those associated with BRCA mutations. We found that PARP1, a key enzyme in DNA repair, was significantly elevated in CML stem cells compared to healthy cells (Figure panel 1). Building on this knowledge, we evaluated MAC681's potential to induce DNA damage (Figure panel 2) and disrupt mitochondrial function (Figure panel 3), leading to cell death. Besides rapid DNA damage induction, MAC681 also degraded PARP1, the essential DNA damage repair component in CML, further enhancing its efficacy against CML. Our in vitro and in vivo experiments confirmed MAC681's ability to reduce tumor growth.
We discovered that MAC681-induced degradation of PARP1 could be prevented by a known PARP1 inhibitor called 3-aminobenzamide (3-ABA). This finding is significant because 3-ABA is commonly used to inhibit PARP1, but our study suggests it may have unexpected chemical interactions with MAC681, potentially reducing its effectiveness. Therefore, caution should be taken when combining these types of compounds.
Furthermore, we found that MAC681-treated leukemia cells released DAMPs, which can enhance the immune response against cancer. Indeed, our experiments in mice showed that MAC681-treated cells could serve as an effective vaccine against CML, significantly reducing tumor growth (Figure panel 4). Additionally, we observed synergistic effects when combined with the novel TKI asciminib in both TKI-sensitive and -resistant models (Figure panel 5).
Overall, our study demonstrates the potential of targeting DNA repair, metabolic pathways, and inducing immunogenic cell death as a promising approach for treating CML. By disrupting key cellular processes and enhancing the immune response, compounds like MAC681 could offer new hope for patients with this challenging disease.
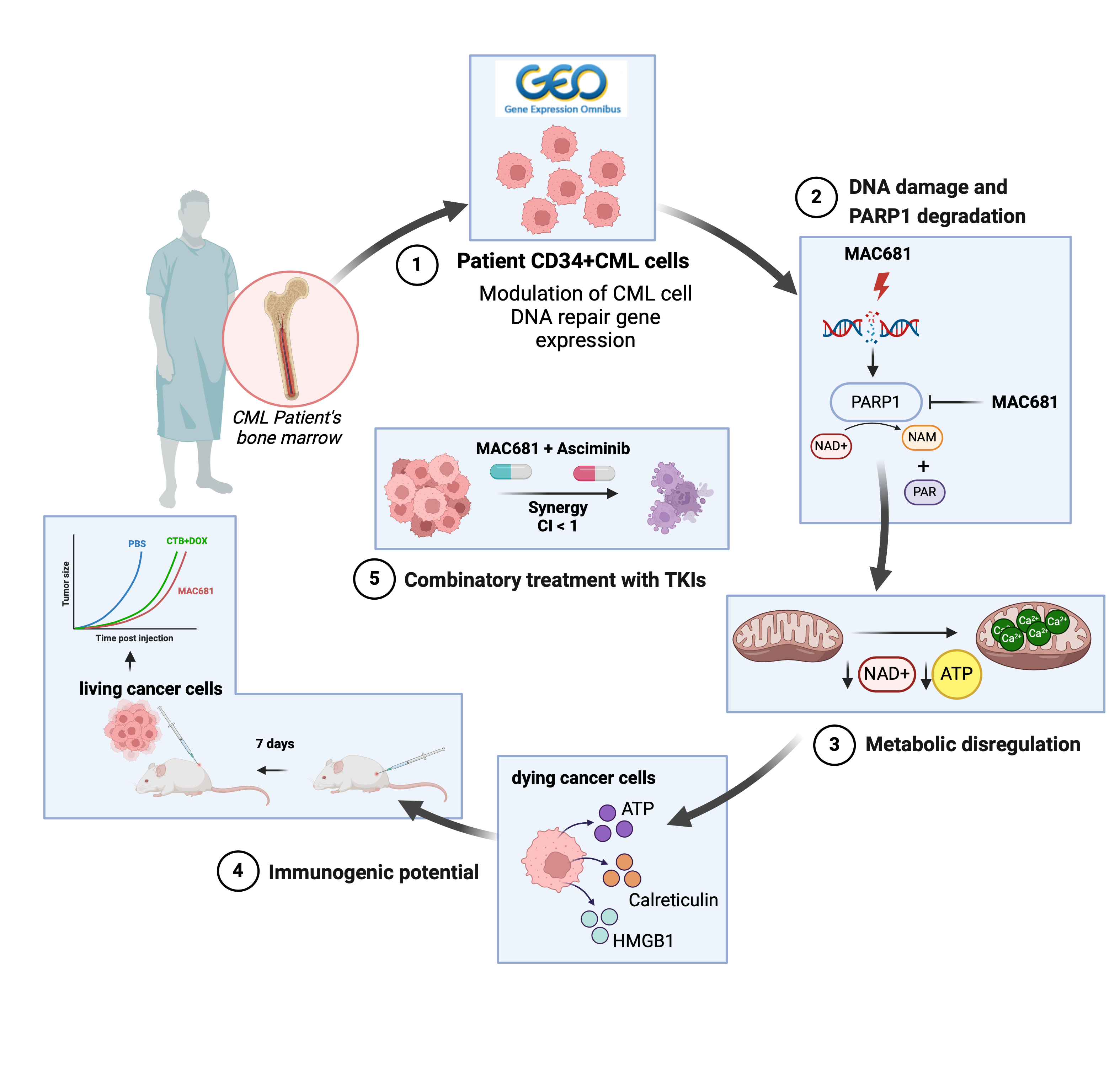 A mechanistic overview describing the effect of methylated indolequinone MAC681 in vitro and in vivo. The figure was created with BioRender.com.
A mechanistic overview describing the effect of methylated indolequinone MAC681 in vitro and in vivo. The figure was created with BioRender.com.
References:
- Eide CA, O'Hare T. Chronic myeloid leukemia: advances in understanding disease biology and mechanisms of resistance to tyrosine kinase inhibitors. Curr Hematol Malig Rep. 2015;10(2):158-66.
- Geelen IGP, Thielen N, Janssen J, Hoogendoorn M, Roosma TJA, Willemsen SP, et al. Treatment outcome in a population-based, 'real-world' cohort of patients with chronic myeloid leukemia. Haematologica. 2017;102(11):1842-9.
- Kuntz EM, Baquero P, Michie AM, Dunn K, Tardito S, Holyoake TL, et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med. 2017;23(10):1234-40.
Follow the Topic
-
Biomarker Research

This is an open access, peer-reviewed journal that encompasses all aspects of biomarker investigation.
Related Collections
With Collections, you can get published faster and increase your visibility.
Oncobiome
This collection of papers delves into the burgeoning field of oncobiome research, exploring the intricate relationship between cancer and the microbiome. The oncobiome encompasses the diverse microbial communities residing in and on the human body, which influence cancer development, progression, and treatment responses. By examining these interactions, our aim is to unravel the complex mechanisms through which the microbiome impacts oncogenesis and therapeutic outcomes.
This compilation highlights cutting-edge research, offering insights into potential diagnostic markers and novel therapeutic strategies, thereby advancing our understanding of cancer biology and paving the way for innovative, microbiome-targeted cancer treatments.
This collection has been peer reviewed by the Editorial Boards of Biomarker Research
Publishing Model: Open Access
Deadline: Ongoing



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in