The birds and the bees: Bacterial sex and its implications in spreading antibiotic resistance
Published in Microbiology
Horizontal gene transfer, or HGT, is the notion that bacteria can share genes laterally amongst themselves, and is about as fascinating and pertinent a topic as any in microbiology. Comparing different bacterial species, each about 1/1000th the size of a grain of sand, might seem silly, especially when comparing larger species like humans and birds. How different can single-celled organisms really be? Well, HGT between distinct bacterial species is analogous to me standing next to an eagle, and growing wings the next day. As crazy (and potentially awesome) as this might seem, bacteria readily undergo HGT, often involving genes that confer antibiotic resistance. For this reason, HGT, and specifically conjugation (a type of HGT), is a major player in the worldwide spread and threat of superbugs.
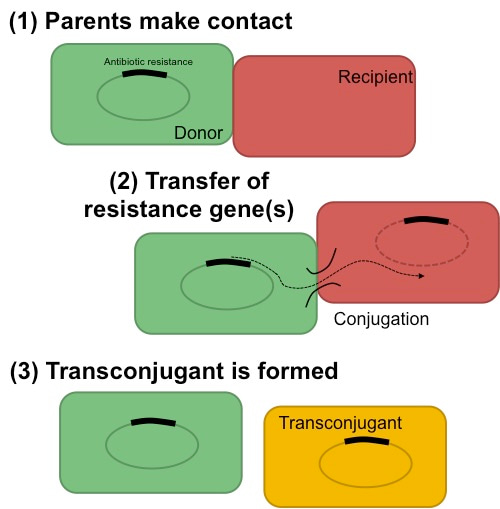
Conjugation process between donor and recipient occurs through direct cell-cell contact, and results in a transconjugant.
When we set out to explore the scientific applications of HGT, we were focused on harnessing the unique aspects of gene transfer towards engineering an evolvable microbial community. However, delving into the literature raised many unanswered questions about the underlying mechanisms of HGT itself; most striking was the lack of a definitive answer to a very fundamental question: do antibiotics actually cause HGT to spread resistance?
In fact, the use of antibiotics has long been implicated in the rise and spread of antibiotic resistance, and in certain cases, the link between antibiotics and conjugation is well documented for antibiotic-induced conjugation – what about for all the other cases where conjugation machinery is already being expressed? The answer to this question has significant clinical implications; after all, if we can predict how antibiotics influence the transfer of resistance, we may be able to develop solutions to prevent it. And so, our research direction shifted.
In our study, we quantified the conjugation efficiency, or the probability of conjugation occurring between a pair of donor and recipient cells. Using a synthetic system, we surprisingly saw that antibiotics did not increase the conjugation efficiency in the absence of selection, in contrast to conventional wisdom. Using a host of clinically relevant pathogens and systems also showed no significant increase in conjugation efficiency, to our surprise (and as synthetic biologists, delight). As is so common with science, the answer to this question led to another. If antibiotics don’t change the conjugation rate, what can explain the increase in transconjugants (resistant bugs resulting from conjugation)?
With a touch of modeling and a bunch of microfluidic experiments, we saw that antibiotic selection can account for the rise of transconjugants. Surprisingly, though, this didn’t mean that more antibiotics always led to more HGT. Instead, we found that antibiotic use often has opposing effects. On one hand, antibiotics suppress the emergence of transconjugants by reduction in one or both of the parents. On the opposing hand, antibiotics promote transconjugants by offering a selective advantage. One effect negates the other, and only together can we determine whether transconjugants will be favored. While intuitive, these results were not obvious.
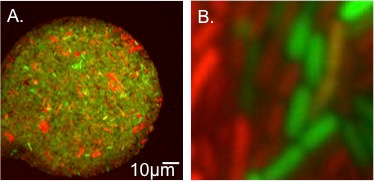
A. Representative microfluidic chamber filled with transconjugants. Transconjugants appear yellow by eye due expression of both mCherry and GFP. B. Close-up view of transconjugants, donors, and recipients.
It’s our hope these results shed light into the dynamics of antibiotic-treated bacteria undergoing dynamic gene exchange, and present a myriad of new, exciting, and clinically relevant scenarios that can be leveraged to stem the spread of resistance worldwide. For example, the potential to help design smarter antibiotic dosing strategies aimed at minimizing the spread of resistance during treatment would be an important step forward against the war on superbugs.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in