The Fibre Fix: The Role of Dietary Fibre in Directing Production of Tryptophan Metabolites by Gut Microbes!
Published in Microbiology

The gut microbiota is crucial in maintaining good health and preventing diseases. It is well known that gut microbial metabolism of the aromatic amino acid tryptophan leads to the production of indole, a precursor of the uremic toxin indoxyl sulfate (IS) that contributes to the pathogenesis of chronic kidney disease (CKD), and its sequelae such as cardiovascular diseases[1,2]. On the other hand, tryptophan-derived microbial metabolites such as indolelactic acid (ILA) and indole propionic acid (IPA), are beneficial for health by having anti-inflammatory, immunomodulatory, and neuroprotective effects [3]. However, it has so far remained unclear what determines whether tryptophan will be converted into indole or into ILA and IPA.
Before starting this project, Tine Rask Licht’s lab at the National Food Institute, Technical University of Denmark (DTU) had developed an interest in the role of gut microbial tryptophan metabolites in health and diseases [3]. Work in the lab headed by (at that time Postdocs) Martin Laursen and Henrik Roager along with other excellent scientists, revealed how ILA is produced by breastfeeding-associated Bifidobacterium species in the infant gut and contributes to immune maturation in early life [4]. During this exciting time, Tine was awarded a Challenge Grant from the Novo Nordisk Foundation to work on the PRIMA project (towards Personalized dietary Recommendations based on the Interaction between diet, Microbiome, and Abiotic conditions in the gut), a collaboration among others with the Roager lab at University of Copenhagen [5]. The aim of the PRIMA project was to understand how diet and intestinal abiotic factors affect gut microbial composition and their activities. Anurag Kumar Sinha, a molecular microbiologist, joined the lab as a postdoc (now a tenure track researcher at DTU) in the summer of 2020 to work on this project. Here, Anurag and Martin (now an associate professor at DTU) teamed up to unravel what controls the metabolic fates of tryptophan in the gut beyond infancy.

Anurag Sinha (left) and Martin Laursen (right)
In essence, we wanted to understand the factors that determine whether beneficial or harmful tryptophan metabolites are produced in the adult gut. Anurag started culturing two model gut bacteria namely Clostridium sporogenes and Peptostreptococcus anaerobius, that are known to perform reductive Stickland fermentation to produce ILA and IPA [6](Fig. 1a). Stickland fermentation of amino acids is a coupled chemical reaction through which clostridium species obtain their energy where oxidative metabolism of one amino acid generates ATP via substrate-level phosphorylation, while the redox balance is maintained by reducing another amino acid [7]. We quantified metabolites by ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) with the help of an experienced chemist, Mikael Pedersen. Interestingly, we found that the Stickland fermentation products of tryptophan were directly proportional to the substrate availability, i.e., tryptophan present in the growth medium (Fig. 1b). The result was also confirmed with the in vitro cultures of human infant fecal cultures.
a
 b
b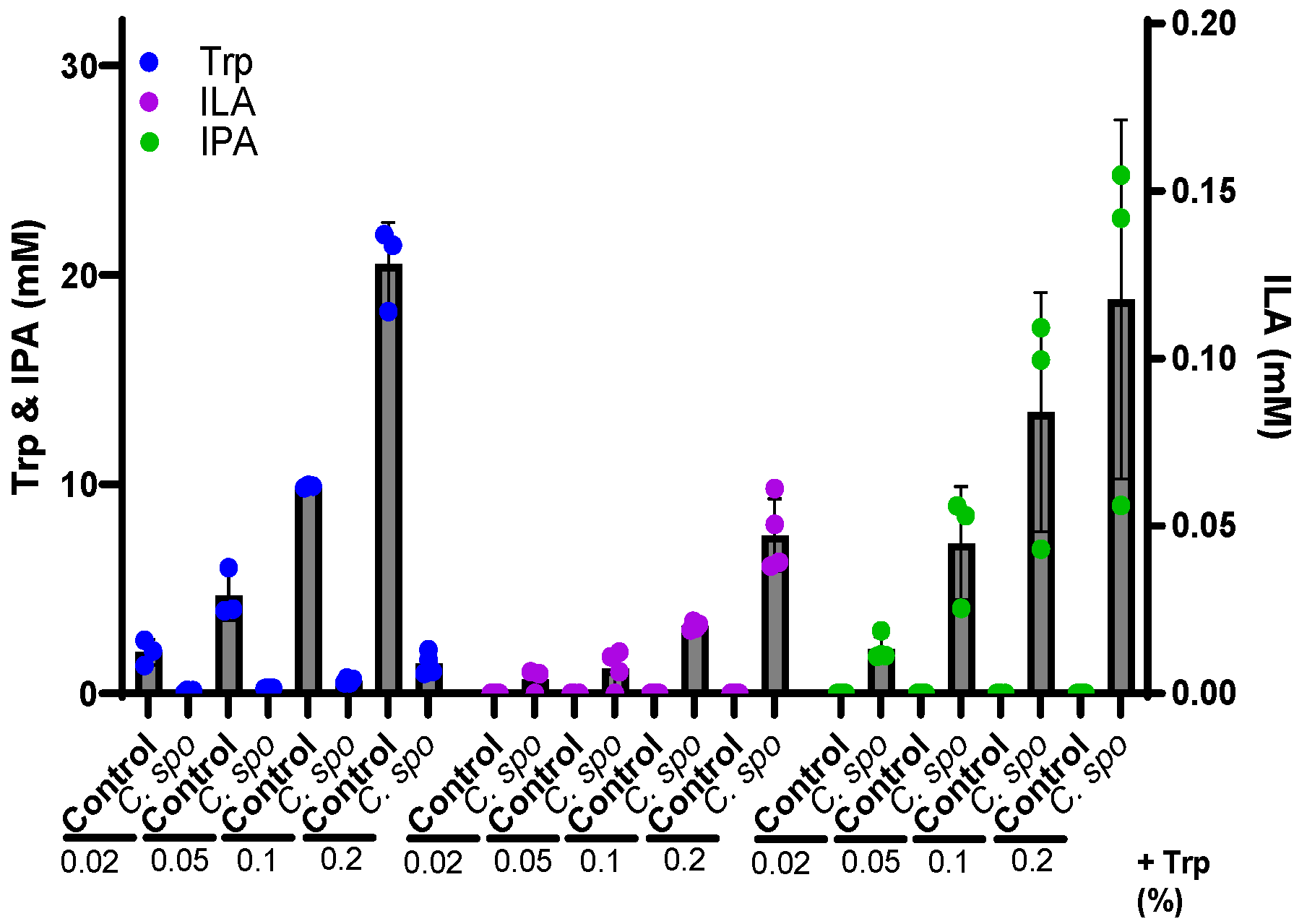
Fig. 1 (a) Stickland fermentation pathway (b) Amount of tryptophan metabolites produced by C. sporogenes in tryptophan-supplemented growth medium.
The next obvious question that came to our mind was how we could make more tryptophan available to ILA, IPA-producing bacteria in the gut. Since indole is the most abundant tryptophan metabolite present in the human and mouse gut, contributing to 50-75% of the total tryptophan metabolites [8], we hypothesized that reducing indole production could redirect tryptophan to other bacteria, leading to increased production of ILA and IPA.
Eureka moment
Indole is produced through a single enzymatic process catalyzed by the tnaA gene-encoded tryptophanase enzyme, which hydrolyses tryptophan into indole, pyruvate, and ammonia. Though many gut bacteria possess the tnaA gene and can produce indole; Escherichia coli (E. coli) is specifically enriched in CKD and responsible for high indole accumulation in CKD patients [9]. In E. coli, the tnaA gene is under the control of carbon catabolite repression, and its expression is thus inhibited by the presence of simple carbohydrates such as glucose, arabinose and pyruvate [10]. Anurag was joined by Julius, a new PhD student in the group, to culture fecal samples in the presence of simple carbohydrates (glucose, maltose and cellobiose). The results were satisfying as simple sugars inhibited indole production by gut bacteria, increasing tryptophan availability and allowing other bacteria to convert tryptophan into ILA. Here we go, our hypothesis proved to be correct!
But simple carbohydrates from the diet do not reach the colon and are unavailable to the gut microbes. How do gut microbes get simple carbohydrates? While we were working on this project, an excellent paper was published that suggested that Bacteroides thetaiotaomicron, a fibre-degrading gut bacterium, can extract mono- and oligosaccharides from dietary fibres and mucus glycans, and cross-feed to other gut bacteria [11]. Our enthusiasm doubled and we felt like we got the answer!
We proposed another hypothesis: Fiber-degrading gut bacteria would break down dietary fibers into simple carbohydrates, which would suppress indole production through catabolite repression and increase tryptophan availability for other bacteria to produce beneficial metabolites like ILA and IPA.
Testing of hypothesis
We designed a defined community of Indole-producing Escherichia coli, ILA- and IPA-producing Clostridium sporogenes and fibre-degrading Bacteroides thetaiotaomicron and cultured them in the growth medium supplemented with low and high tryptophan and with and without the dietary fibre pectin. Morten Rybtke, an excellent postdoc (now an assistant professor at the University of Copenhagen), quantified relative gene expression by RT-qPCR and showed that the presence of pectin leads to upregulation of arabinose and xylose utilizing genes and downregulation of tnaA gene in E. coli. Corroborating this, indole was inhibited, and ILA was increased in the presence of pectin, confirming our hypothesis. To validate the findings in vivo, Martin colonized germ-free mice with the same defined community and fed them low and high tryptophan and with and without pectin. The pectin-mediated inhibition of indole was also confirmed in vivo (Fig. 2).
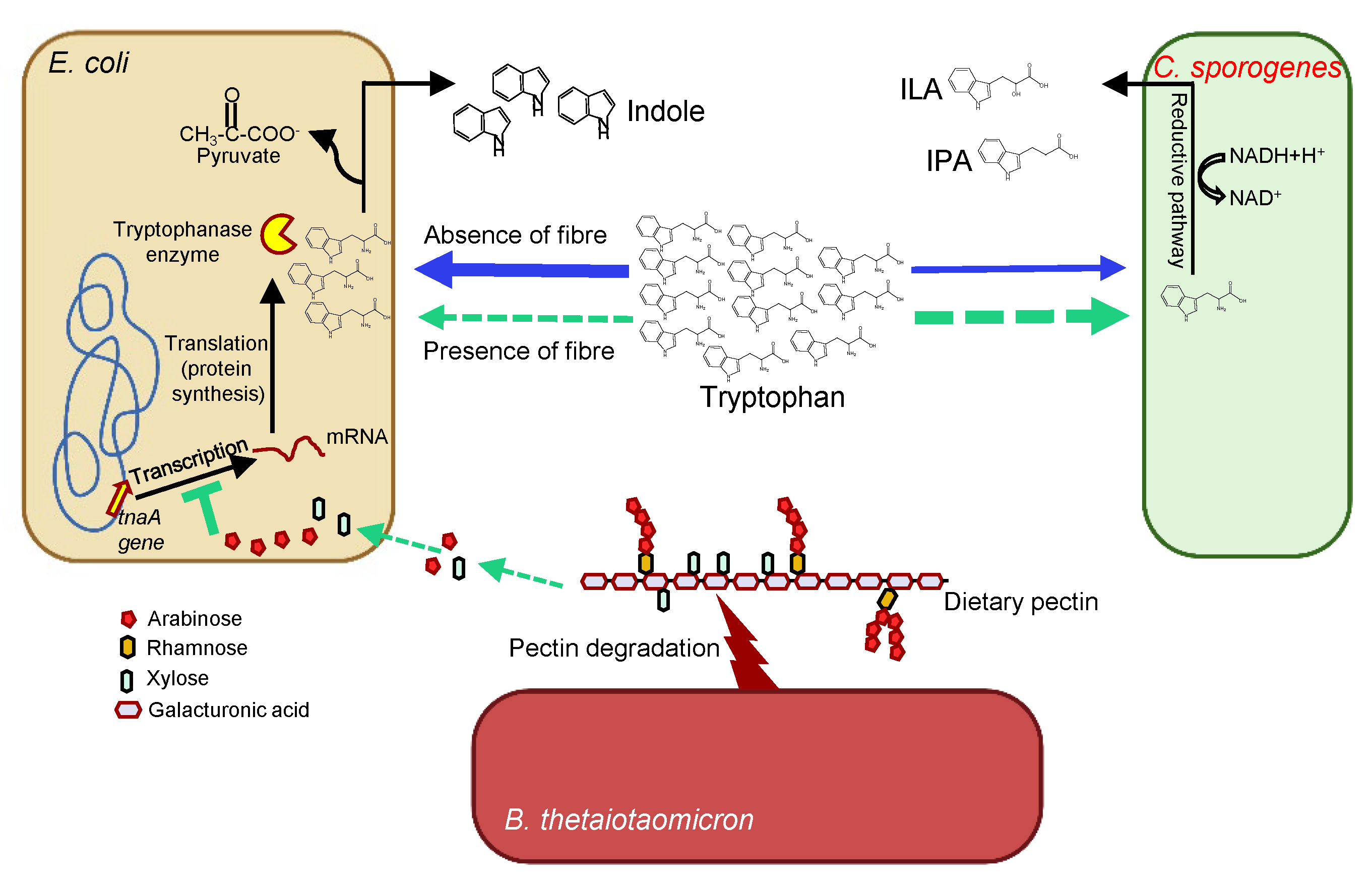
Fig. 2 A model to illustrate the role of Dietary fibre and substrate-mediated impact on tryptophan metabolism.
To further demonstrate that this principle applies to the diverse human gut microbiota, Nicola Procházková (Postdoc in the Roager Lab) provided adult human fecal samples that she collected (and characterized the gut microbiome in) for the human baseline PRIMA study [12] and we performed further in vitro and gnotobiotic in vivo studies, together with Anna Pii Hjørne (PhD student at DTU), showing that fibre-mediated control of tryptophan metabolism and inhibition of indole production is a common phenomenon in diverse gut microbial communities.
Our journey
Working on this project was an excellent journey that we thoroughly enjoyed with a team of many outstanding scientists. During the research, we found many unexplained observations in the literature regarding correlations between the consumption of dietary fibres and levels of microbial tryptophan metabolites in feces and blood. We believe that our results provide a rational explanation for these observations. Our study is unique in gut microbiome research because our explanation focuses not on changes in the abundance or composition of gut bacteria but rather on the regulation of their gene expression, which alters the microbial metabolites produced. We believe this study will be a milestone, paving the way for future investigations into the regulatory factors affecting various other gut microbial metabolites.
(The poster image was created with BioRender.com)
References:
- Cheng, T.-H. et al. (2020) Indoxyl Sulfate, a Tubular Toxin, Contributes to the Development of Chronic Kidney Disease. Toxins (Basel). 12, 684
- Hung, S.-C. et al. Indoxyl Sulfate: A Novel Cardiovascular Risk Factor in Chronic Kidney Disease. DOI: 10.1161/JAHA.116.005022
- Roager, H.M. and Licht, T.R. (2018) Microbial tryptophan catabolites in health and disease. Nat. Commun. 9, 1–10
- Laursen, M.F. et al. (2021) Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 6, 1367–1382
- The Foundation awards DKK 60 million for research on the role of gut microbiota in diet and health - Novo Nordisk Fonden[Online]. Available: https://novonordiskfonden.dk/en/news/the-foundation-awards-dkk-60-million-for-research-on-the-role-of-gut-microbiota-in-diet-and-health/. [Accessed: 08-Jun-2024]
- Dodd, D. et al. (2017) A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652
- LEONARD HUBERT STICKLAND (1934) Studies of the metabolism of the strict anaerobes (genus: Clostridium ). Biochem. J. 28, 1746–59
- Darkoh, C. et al. (2015) A rapid and specific method for the detection of indole in complex biological samples. Appl. Environ. Microbiol. 81, 8093–8097
- Lobel, L. et al. (2020) Diet posttranslationally modifies the mouse gut microbial proteome to modulate renal function. Science (80-. ). 369, 1518–1524
- Li, G. and Young, K.D. (2014) A cAMP-independent carbohydrate-driven mechanism inhibits tnaA expression and TnaA enzyme activity in Escherichia coli. Microbiol. (United Kingdom) 160, 2079–2088
- Schmidt, F. et al. (2022) Noninvasive assessment of gut function using transcriptional recording sentinel cells. Science (80-. ). 376, eabm6038
- Procházková, N. et al. Gut environmental factors explain variations in the gut microbiome composition and metabolism within and between healthy adults. DOI: 10.1101/2024.01.23.574598
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in