The ‘forever-young’ secret of scallop sex chromosomes: exceptions to the ‘general’ rules
Published in Ecology & Evolution

Sexual reproduction is one of the most widespread and fascinating biological phenomena. The evolution of sex chromosome is among the core issues of evolutionary biology. Since more than 100 years ago, the classic theory has been assuming sex chromosomes that originated from autosomes will eventually become heteromorphic through recombination suppression and gradual chromosomal degeneration (Figure 1). This theory dominates the basic concepts of almost all sex-related studies in the field, and heteromorphic sex chromosomes are often thought to be the "ultimate-fate" of the evolutionary pathway of sex chromosomes1,2,3. However, contrary to this prediction, there is growing evidence that species with homomorphic sex chromosomes are curiously prevalent in the animal kingdom2. To explain this paradox, researchers still hold with the classical theory, assuming all the homomorphic sex chromosomes are not old enough or sometimes ignore them as "exceptions to the rules"4. 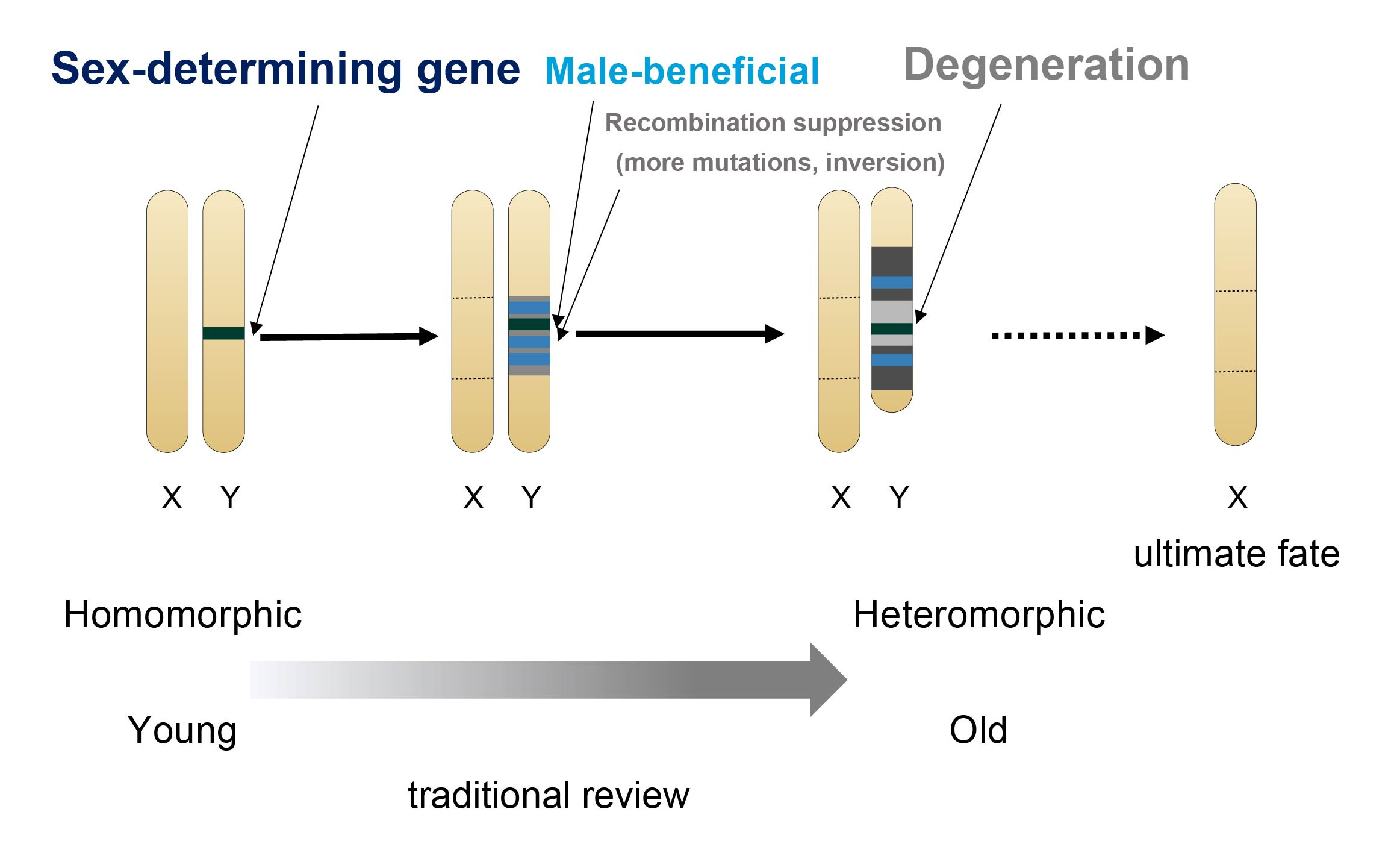
Figure 1. Classical model of sex chromosome evolution (modified from ref3).
Yet, could there be a different answer? In the summer of 2019, I joined Wang's lab at Sars-Fang Centre, Ocean University of China and study a fairly related project for my PhD dissertation. Our group has long been working on molluscan sex development and regulation – a research direction led by Prof. Lingling Zhang. At the time, we noticed that although molluscs constitute the second largest and Precambrian-originated animal phylum, their ancient homomorphic sex chromosomes and sex-determining genes had not been characterized largely due to the challenge of identifying homomorphic sex chromosomes. Our initial goal was to improve the genome and identify the sex chromosome in Yesso scallop. Under the guidance of Prof. Lingling Zhang, we assembled the chromosome-level and haplotype-solved genome of Yesso scallop, which allowed us to identify a W-specific region which was only 38kb long. We were excited to see a candidate sex-determining gene-FOXL2 (which has long been suspected as a key sexual regulator in bivalve molluscs by our and other groups5,6) in the downstream of the specific region, and I was then preparing to write an article on the first report of homomorphic sex chromosome and sex-determining gene in Lophotrochozoa.
Then, the first turning point of this project came, from a daily discussion with Prof. Shi Wang. Considering that Yesso scallop is a basal-branch-derived species with a slow-evolving or "fossil-like" genome7(https://ecoevocommunity.nature.com/posts/15927), he incisively discovered that this would be an excellent material for studying the evolution of ancient homomorphic chromosomes. To obtain a macroscopic view of the evolution of scallop sex chromosomes, we assembled and collected the chromosome-level genomes of eight representative species across the entire Pectinidae family by integrating third-generation sequencing and Hi-C technology. By carefully screening for genome-wide sexual SNPs and sex-specific regions, we successfully identified sex-determining genes in four gonochoristic scallops (FOXL2 for Yesso scallop and Chinese scallop8; ZNF226l for Sea scallop and CYP3A24l for Japanese moon scallop). After phylogenetic analysis and sex-specific repeat sequence dating, it was striking to see the undifferentiated state of sex chromosomes in Yesso scallop which has been maintained for at least 350 million years. It is the oldest known homomorphic chromosome in the animal kingdom, even as twice long as the ones in birds and mammals. This unexpected finding strongly refutes the traditional assertion that homomorphic sex chromosomes are still too young.
So, how could scallop sex chromosomes remain undifferentiated for 350 million years? Answering this question not only helps to understand the maintenance mechanism of homomorphic sex chromosomes, but also fills up a knowledge gap on the evolutionary puzzle of sex chromosomes from homomorphic to heteromorphic. We innovatively adopted a dynamic analytical strategy to profile gonad developmental transcriptome, and found a large number of reversible sex-biased genes (rSBGs) on homomorphic chromosomes (sex preference switches between males and females at different stages of gonad development), while only few on heteromorphic sex chromosomes. By comparing the genome distribution and expression patterns of rSBGs in scallops, fruit fly, fish, birds, human, etc., we revealed that rSBGs could play a “barrier” role to restrict the expansion of the recombination suppression region of sex chromosomes and thus maintain the undifferentiated status of sex chromosomes. It clarified that the process of sex chromosomes from homomorphic to heteromorphic requires the drive of sexual dimorphism and sexual selection pressure, and the sex conflicts of rSBGs could be resolved by subfunctionalization after gene doubling. Our findings lead to a novel evolutionary model for homomorphic and heteromorphic sex chromosomes (Figure 2).
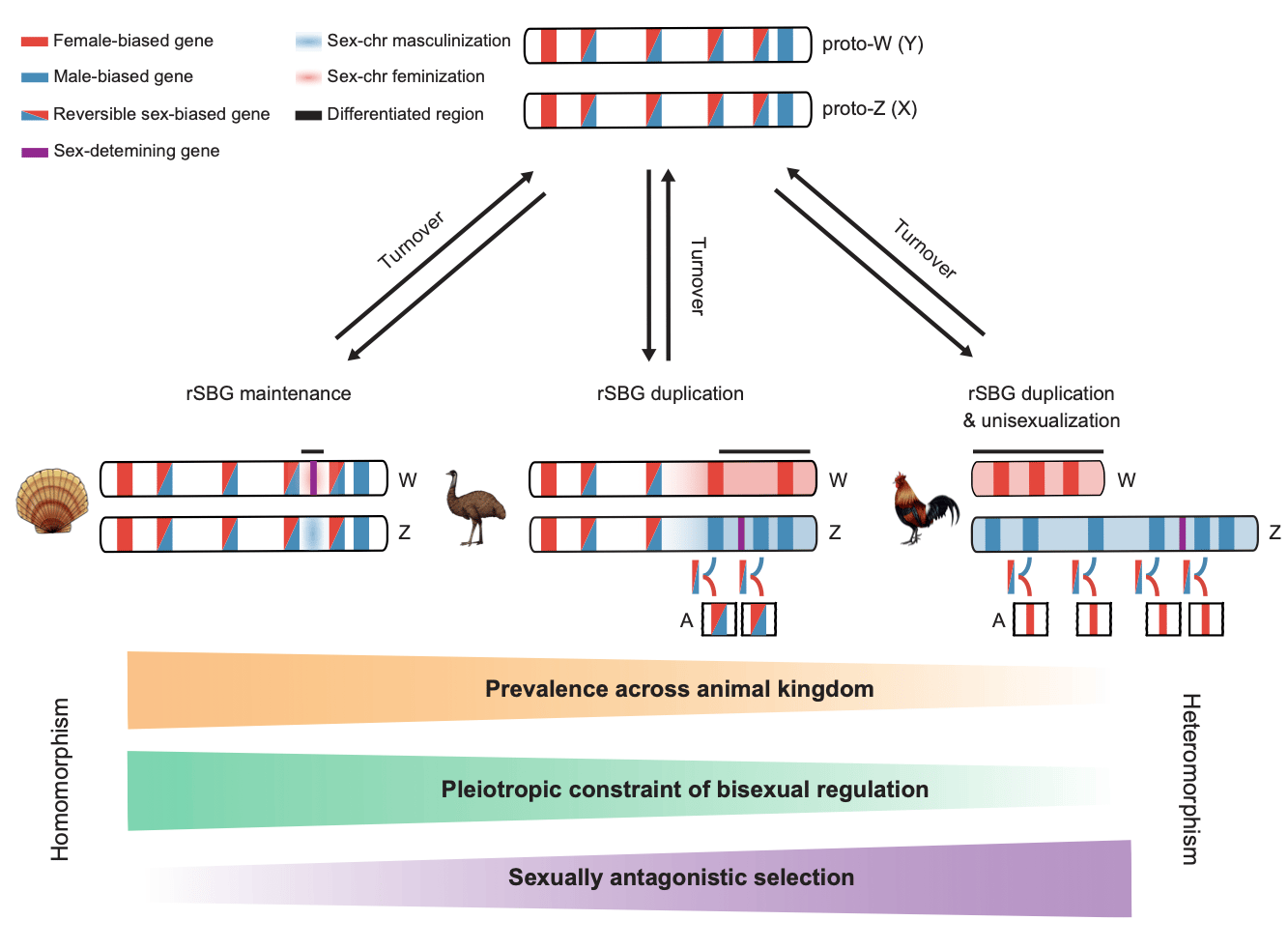
Figure 2. A new model for sex chromosome evolution.
We also noticed the sex-determining genes of Sea scallop and Japanese moon scallop are different than the ones in Yesso scallop and Chinese scallop, how did they change their sex-determining genes? Classic theory holds that a new sex-determining gene needs to compete with and replace the old one, yet this model does not well explain how the old sex-determining genes are handed over. When we were very confused and struggled for a long time to understand the connections among those sex-determining genes, we invited Dr. Kun Wang at Northwestern Polytechnical University to give a report. I never expected the second turning point of the project came, when he mentioned that conserved non-coding elements (CNE) play important roles in the evolution of lungfish. That inspired us the connections we were seeking may not be on those genes themselves, but on their regulatory elements. With the help of Dr. Lisui Bao, we found there’re indeed CNEs in the sex-link regions among those species. They exist specifically upstream of the sex-determining genes and putting together with the consistent findings of our parallel project (including ATAC-seq and dual-luciferase reporter assay by the contributions of labmates) provides strong functional evidence for this region containing a common translocatable sex-determining enhancer. Thus, we propose an elegant model of sex-determining gene turnover in scallop, where no competition is required and sex chromosome transitions are mainly through the translocation of W-related sex-determining enhancer to achieve. We call it “inheritance model”, as the model nicely explains the birth of new sex-determining genes and the demise of old sex-determining genes.
The discoveries in this project reveal that the maintenance of homomorphic sex chromosomes should be considered as the "regular rule" of sex chromosome evolution, and the differentiation of heteromorphic sex chromosomes requires additional evolutionary driven force, which could represent a branch on the entire evolutionary pathway rather than "ultimate-fate".
References
- Charlesworth, B. The evolution of sex chromosomes. Science 251, 1030–1033 (1991).
- Wright, A. E., Dean, R., Zimmer, F. & Mank, J. E. How to make a sex chromosome. Nat. Commun. 7, 12087 (2016).
- Vicoso, B. Molecular and evolutionary dynamics of animal sex-chromosome turnover. Nat. Ecol. Evol. 3, 1632–1641 (2019).
- Furman, B. L. S. et al. Sex chromosome evolution: so many exceptions to the rules. Genome Biol Evol. 12, 750–763 (2020).
- Li, R. et al. FOXL2 and DMRT1L are Yin and Yang genes for determining timing of sex differentiation in the bivalve mollusk Patinopecten yessoensis. Front. Physiol. 9, 1166 (2018).
- Zhang, N., Xu, F. & Guo, X. Genomic analysis of the Pacific oyster (Crassostrea gigas) reveals possible conservation of vertebrate sex determination in a mollusc. G3 4, 2207–2217 (2014).
- Wang, S. et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1, 120 (2017).
- Li, Y. et al. Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat. Commun. 8, 1721 (2017).
Follow the Topic
-
Nature Ecology & Evolution

This journal is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biodiversity and ecosystem functioning of global peatlands
Publishing Model: Hybrid
Deadline: Jul 27, 2026
Understanding species redistributions under global climate change
Publishing Model: Hybrid
Deadline: Jun 30, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in