The mysterious origin of metazoan larvae: Larva-first or Adult-first?
Published in Ecology & Evolution

How diverse metazoan primary larvae originated? That is a fascinating but also a challenging question, which have intrigued immense research interests and debates and puzzled researchers for more than a century. In a Nature Ecology & Evolution paper, we address this question by comprehensive evolutionary analyses of the ontogenetic transcriptomes of molluscs and other major metazoan groups.
This project was started when I was a PhD student. Our research group has been working on scallops as research animals for decades. After accomplishment of the first high-quality reference genome for the scallop Patinopecten yessoensis in 20171, we decided to generate a comprehensive scallop transcriptome atlas, aiming to gain deep insights of spatial and temporal expression patterns of scallop genes for developmental regulation and evolution. At that time, we had no thoughts of relating the project to larval origin and evolution.
The turning point of the project came from an ordinary discussion between me and Prof. Shi Wang. When I showed a heatmap about the transcriptome similarity among all scallop ontogenetic stages to him, he incisively found that scallop trochophore seemed to be a distinct larval stage with unexpected transcriptome dissimilarity to its adjacent stages (Figure 1). When looking deeper, we found that the trochophore stage had an outstanding ability of incorporating new genes, which accounts for its rapid transcriptome evolution as well as being a “disruptive” stage among the whole life cycle of scallop (Figure 1). We were very excited as these findings directly argue against the prevailing opinion about the larval origin (i.e., “larva-first” hypothesis, see below).
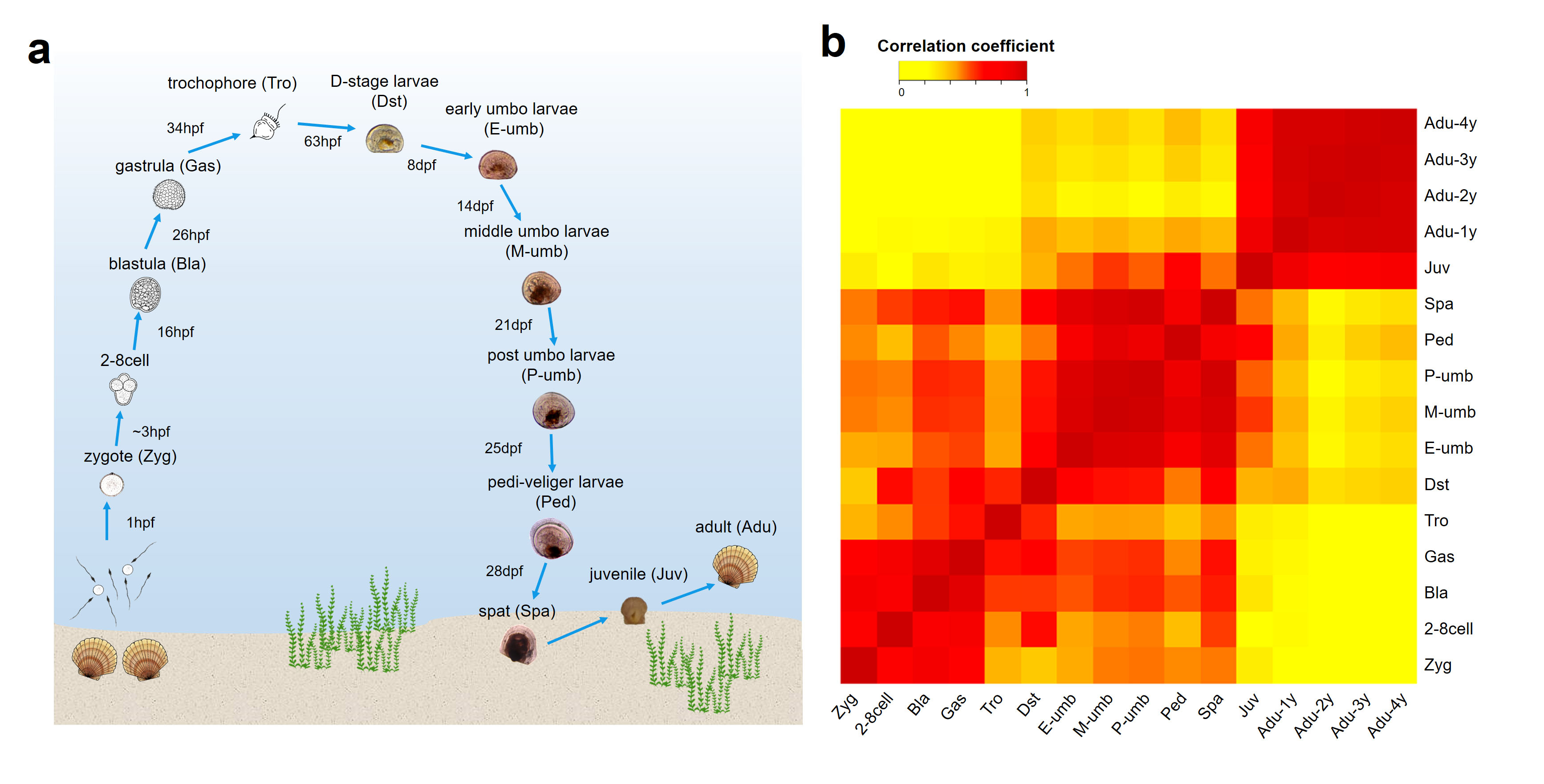
Figure 1. Comparison of transcriptome similarity across scallop ontogeny.
(a) The life cycle of scallop P. yessoensis. (b) Transcriptome comparison among 16 ontogenetic stages.
In light of these findings, we decided to put a focus on resolving the evolutionary origin of metazoan larvae, which is currently among major enigmas in animal evolution and biology2-4. There are two main viewpoints dominate the long history of debate about the origin and evolution of metazoan larvae: the prevailing “larva-first” hypothesis5 and the opposing “adult-first” hypothesis6. The latter seems counterintuitive as it challenges the general belief (i.e., developmental progression mimicking the evolutionary history). Historically, much of the long-standing debate had focused on larval morphology and developmental cell biology, with less evidence from molecular-based studies. With the rapid development of advanced sequencing technologies, omics-scale investigations would provide an excellent opportunity for investigating the evolution origin of metazoan larvae. At least 15 metazoan phyla possess a transient larva-bearing biphasic life cycle. To address our fundamental question, we realized that our omics-scale analyses should be expanded to all the major metazoan lineages with primary larval stages.
Despite the rapid increasing of genome sequencing projects for diverse metazoan lineages, full sets of transcriptomic resources spanning the entire animal ontogeny remain scarce, which is however crucial for our project. We wish to thank Prof. Caihuan Ke, Prof. Bo Dong and Prof. Zhifeng Zhang for kindly sharing their ontogenetic transcriptome data of abalone, sea squirt and spoon worm to us. We finally obtained more than 200 transcriptome data from eight metazoan species. We also constructed a custom protein database that containing 97,308,110 protein sequences of 806,786 species from NCBI Protein database for gene age estimation, which is crucial for the transcriptome age-based analyses in our study. It is not an easy job to handle such big dataset, we had worked days and nights to make the project moving forward. But as difficult as the process became, it was always twice as much fun and definitely worth our efforts.
Finally, our transcriptome-age analysis revealed the wide presence of young larval transcriptomes in major metazoan lineages, providing strong evidence against the prevailing “larva-first” hypothesis. However, different from current “adult-first” model that assumed multiple, independent larval intercalations in major animal lineages, our findings support an “adult-first” evolutionary scenario of single metazoan larval intercalation and the first appearance of proto-larva likely occurred after the divergence of direct-developing Ctenophora from metazoan ancestor (Figure 2). We also revealed that cell signaling/communication genes (e.g., caveolin and innexin) were likely crucial for larval evolution, and proposed an evolutionary scenario for the emergence of a pelagic larval stage through evolutionary addition of the ‘larval’ features/portion (e.g., caveolin/innexin-expressing domain) to the existing ‘adult’ portion (e.g., ATP1B-expressing domain).
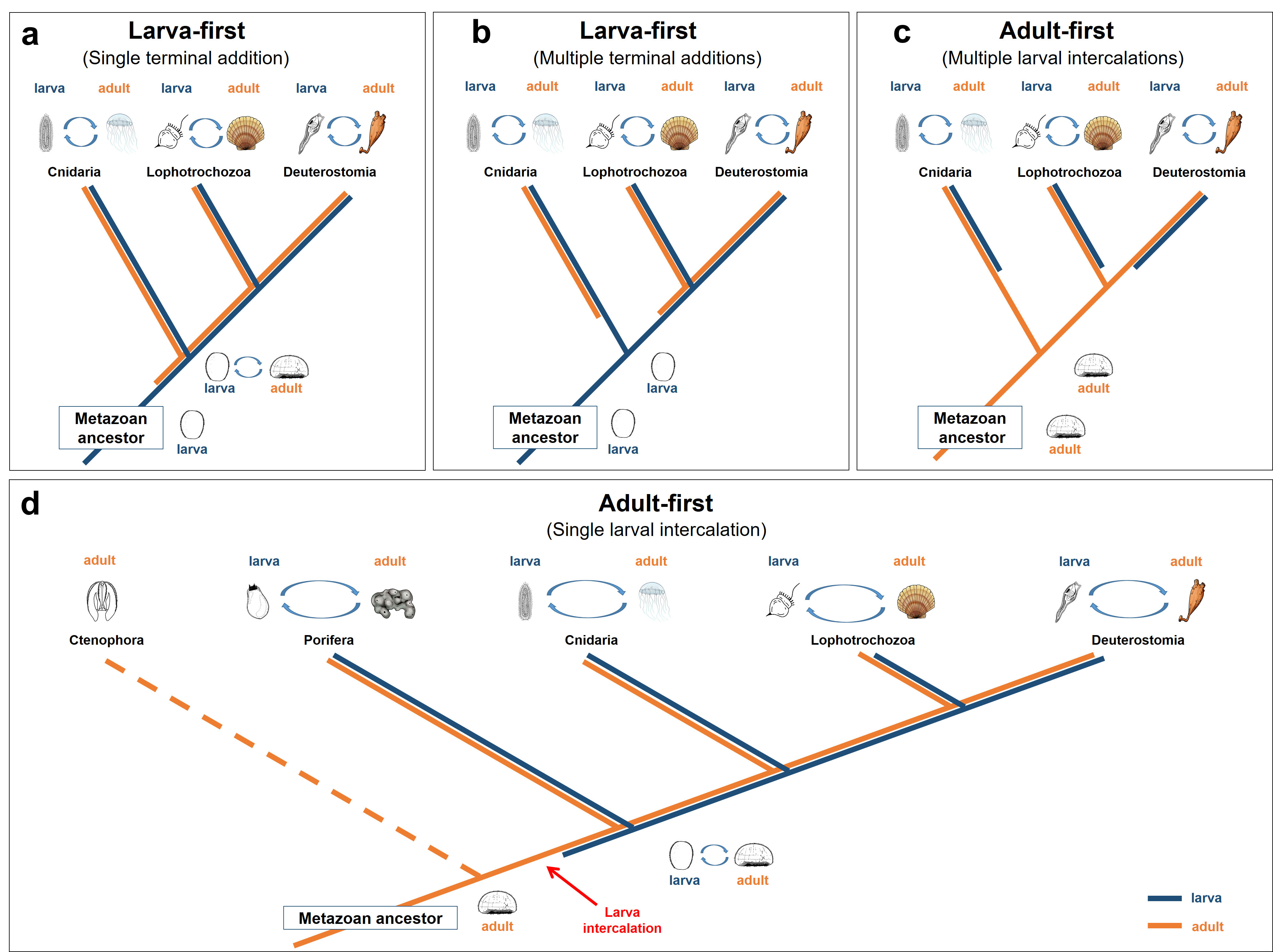
Figure 2. Different evolutionary scenarios for the evolution of metazoan biphasic life cycle.
(a-c) Summary of current main models of “larva-first” and “adult-first” theory according to Marlow4. (d) The proposed “adult-first” model by this study that assumed the single intercalation origin of metazoan larvae.
When looking back, the project would have been totally different without Prof. Wang’s acute sense of the “disruptive” trochophore stage. I am very grateful to him for giving me constant guidance and confidence throughout the project. Thanks also to each team member for their contribution to keeping the project moving on in their area of expertise.
Proposing a new way of thinking about the larval origin and evolution is of course not the end of our story. Instead, our study invokes many follow-up questions, e.g., How new genes were originated and incorporated into the larval stage? What are the functions of new genes in larval development and evolution? How the conserved larval/adult marker genes contribute to larval evolution and across-lineage divergence? All these questions are worthy of further investigations in diverse metazoan lineages, which would definitely deepen our understanding of the origin and evolution of metazoan biphasic life cycle.
References
1 Wang, S. et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1, 0120 (2017).
2 Strathmann, R. R. Hypotheses on the origins of marine larvae. Ann. Rev. Ecol. Syst. 24, 89-117 (1993).
3 Arenas-Mena, C. Indirect development, transdifferentiation and the macroregulatory evolution of metazoans. Philos. Trans. R. Soc. B 365, 653-669 (2010).
4 Marlow, H. in Evolutionary Ecology of Marine Invertebrate Larvae Ch. 2, 16-33 (Oxford University Press, 2018).
5 Nielsen, C. in Evolutionary Ecology of Marine Invertebrate Larvae Ch. 1, 3-15 (Oxford University Press, 2018).
6 Raff, R. A. Origins of the other metazoan body plans: the evolution of larval forms. Philos. Trans. R. Soc. B 363, 1473-1479 (2008).
Follow the Topic
-
Nature Ecology & Evolution

This journal is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biodiversity and ecosystem functioning of global peatlands
Publishing Model: Hybrid
Deadline: Jul 27, 2026
Understanding species redistributions under global climate change
Publishing Model: Hybrid
Deadline: Jun 30, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in