The story of the enigmatic sponge symbionts Candidatus Nemesobacterales
Published in Microbiology

Sponges (phylum Porifera) are among the most primitive animals and home to dense, diverse and yet distinct microbial communities. Sponge-microbe interactions underpin the evolutionary success of Porifera and represent one of the oldest symbioses between animals and microbes. So far, most studies in the field of sponge microbial ecology have focused on the diversity of the sponge microbiota and only until recently metagenome-based methods have granted access to functional aspects of the sponge microbiome. In this study, we shed light on the phylogeny, metabolism, morphology and localization of a sponge-specific order, Candidatus Nemesobacterales of the Desulfobacterota phylum using genome-resolved metagenomics.
In an earlier work from our lab on the microbial diversity of deep-sea sponges, an OTU classified as an uncultured member of the SBR1093 phylum showed the highest abundance in all individuals of the sponge Geodia barretti, collected from the North Atlantic (Figure 1).
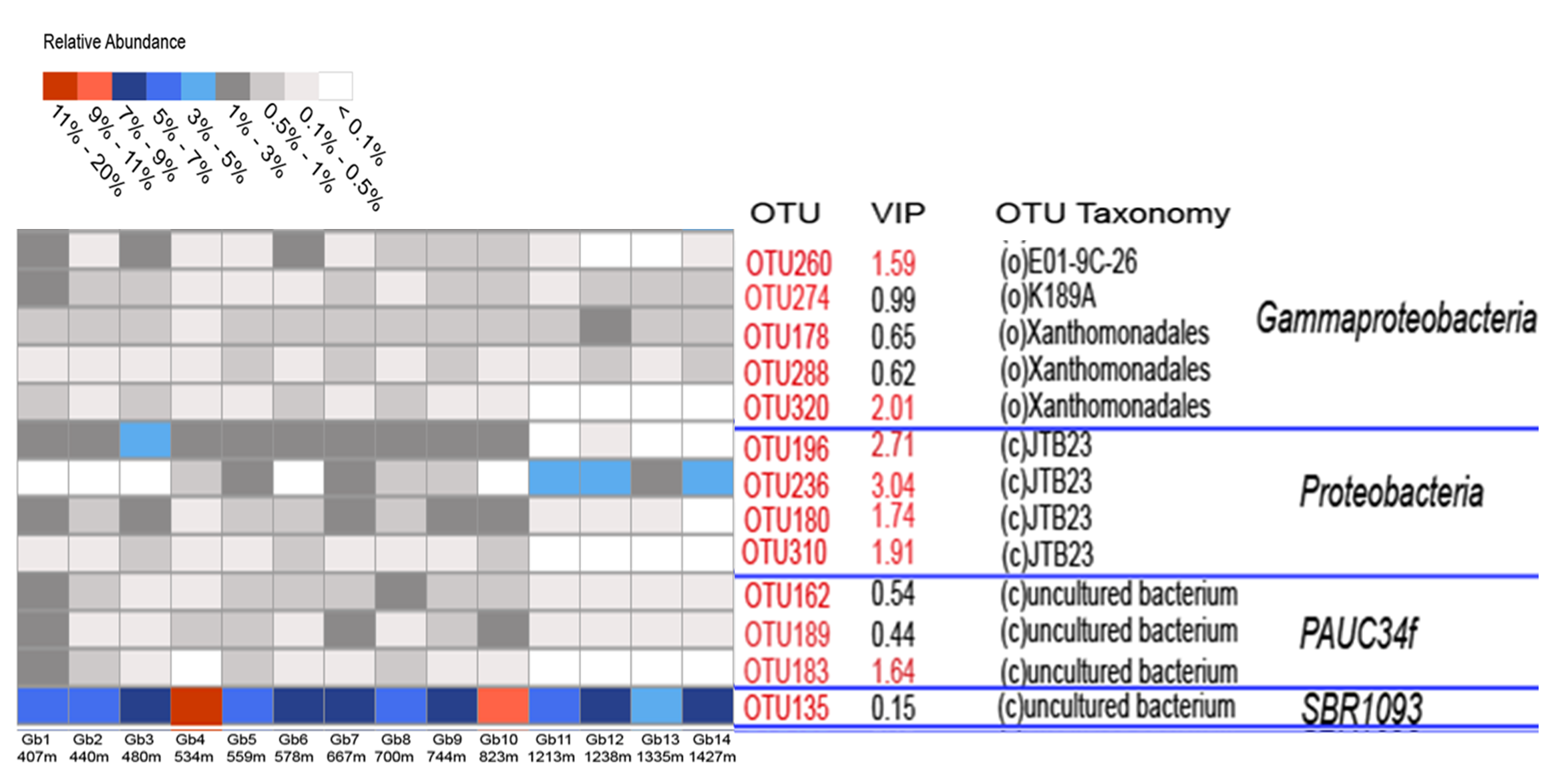
Figure 1. Heatmap showing the relative abundance of certain OTUs in Geodia barretti (Gb) samples from different depths. The OTU with the highest abundance (OTU135) was classified as "uncultured bacterium" of the SBR1093 phylum (unpublished data-source Detmer Sipkema).
A quick literature research revealed that there were only very few studies on SBR1093 and even though they seemed widely distributed, there was no information at all on the role of these bacteria in sponges. This was enough to draw our attention and lead us to a new research adventure!
Asimenia Gavriilidou, with the help of Anastasia Galani and Michelle Schorn joined forces and collected all metagenome-assembled genomes (MAGs) classified as "SBR1093" or "Dadabacteria", found both in-house and in public repositories. First attempts of phylogenomic analysis revealed that there are missing pieces in this puzzle, a yet-unresolved phylogenetic placement in the tree of life.
Our phylogenomic recontruction placed all MAGs coming from sponges in one clade in the Ca. Desulfobacterota phylum uncovering a sponge-specific order, Ca. Nemesobacterales (named and described here) represented by three families, incl. a previously undescribed one. Comparative genomic analysis gave the first insights into their functional repertoire followed by evidence that Ca. Nemesobacterales are metabolically divergent from their seawater counterparts attributed to distinct genomic features suggesting niche adaptation. Most importantly, Burak Avci managed to take the first picture of Ca. Nemesobacterales in the tissue of Geodia barretti using FISH (Banner photo) which revealed their presence both in the mesohyl and inside sponge cells surrounding sponge nuclei.
The story behind the name
Nemesobacterales were named after the ancient greek goddess Nemesis, daughter of Oceanus (Ocean). According to greek mythology, Nemesis was associated with vengeance but also restoring the balance between good and evil by giving rewards for noble acts and punishing for evil ones. We considered this study a reward at last, even though "punishment" came earlier from the sponges which were very difficult to collect during our sampling expeditions. We wouldn't have accomplished the sample collection without the help of the late Hans Tore Rapp (University of Bergen) to whom we dedicate this study.
If you want to know more about the symbiotic lifestyle of Ca. Nemesobacterales, read the full story here.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in