The world's longest selection experiment on mice provides unique animal models for exploring the architecture of polygenic traits
Published in Ecology & Evolution
At the Research Institute for Farm Animal Biology (FBN) in Dummerstorf, Germany, a unique set mouse lines were generated over the course of more than 50 years of artificial selection. These lines have evolved impressive phenotypes of high fertility, body size and endurance fitness. To our knowledge, this is the longest selection experiment ever conducted on mice and we were keen on peering into the genomes of these unique animals, in order to uncover known and new loci associated to the traits under selection.
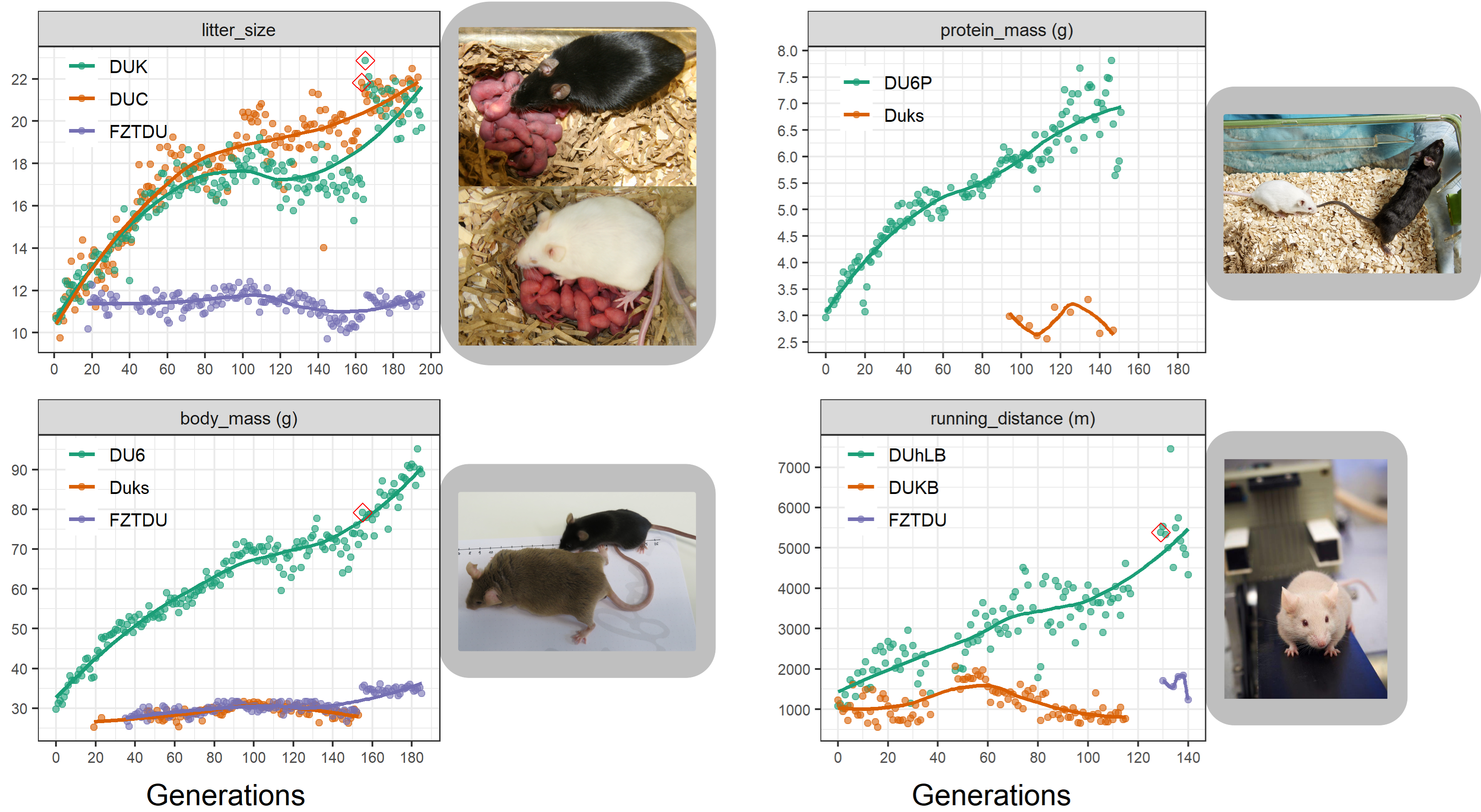
As a population adapts to a selective pressure, the alleles responsible for adaptation become more and more frequent over time. To identify the subtle allele frequency changes underlying complex traits, neutral evolution needs to be rigorously modelled, requiring genetic information not only from the present population but also from ancestral populations, ideally the founders.
Despite the fact that the Dummerstorf mouse lines are outbred mouse populations, we found high levels of inbreeding within each line, resulting from genetic drift after a severe population bottleneck in 2011. Consequently, selected mice are highly homozygous, while at the same time, lines are genetically uniform and distinct from each other.
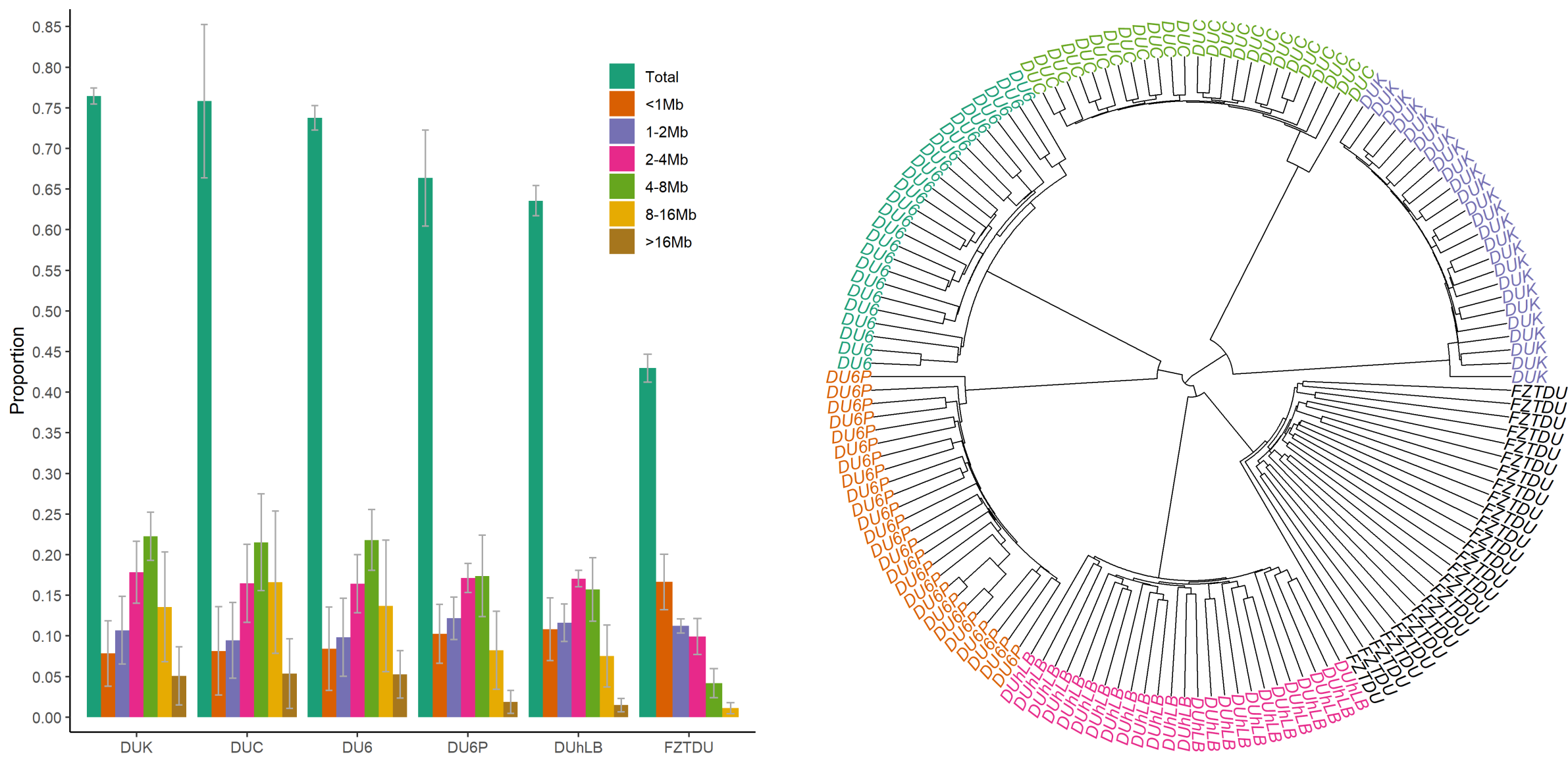
Since genetic information from ancestral generations was not available (this experiment was started in 1969, before the genomic era) and pedigrees were incomplete, detection of signatures of selection was practically impossible. It was also unfeasible to know if response to selection resulted from large- or small-effect alleles, though the complex nature of the selected traits led us to infer that the former is rather unlikely.
The historical resources at our disposal were thus limited, so we tried to come up with an alternative approach that would at least give us an idea of which genes could be involved in the evolution of the selected traits. First, we obtained the genomewide genetic differentiation of each selected line relative to the control line (the control line as a proxy of the founder population, exposed only to genetic drift). We then looked for regions of line-specific high genetic differentiation, each one corresponding to a genomic location in which only the target line is highly differentiated from the control line, while all remaining lines are undifferentiated. This approach revealed a number of candidate genes, some of which are already known to be relevant in the biology of fertility, body mass, muscle growth, and endurance.
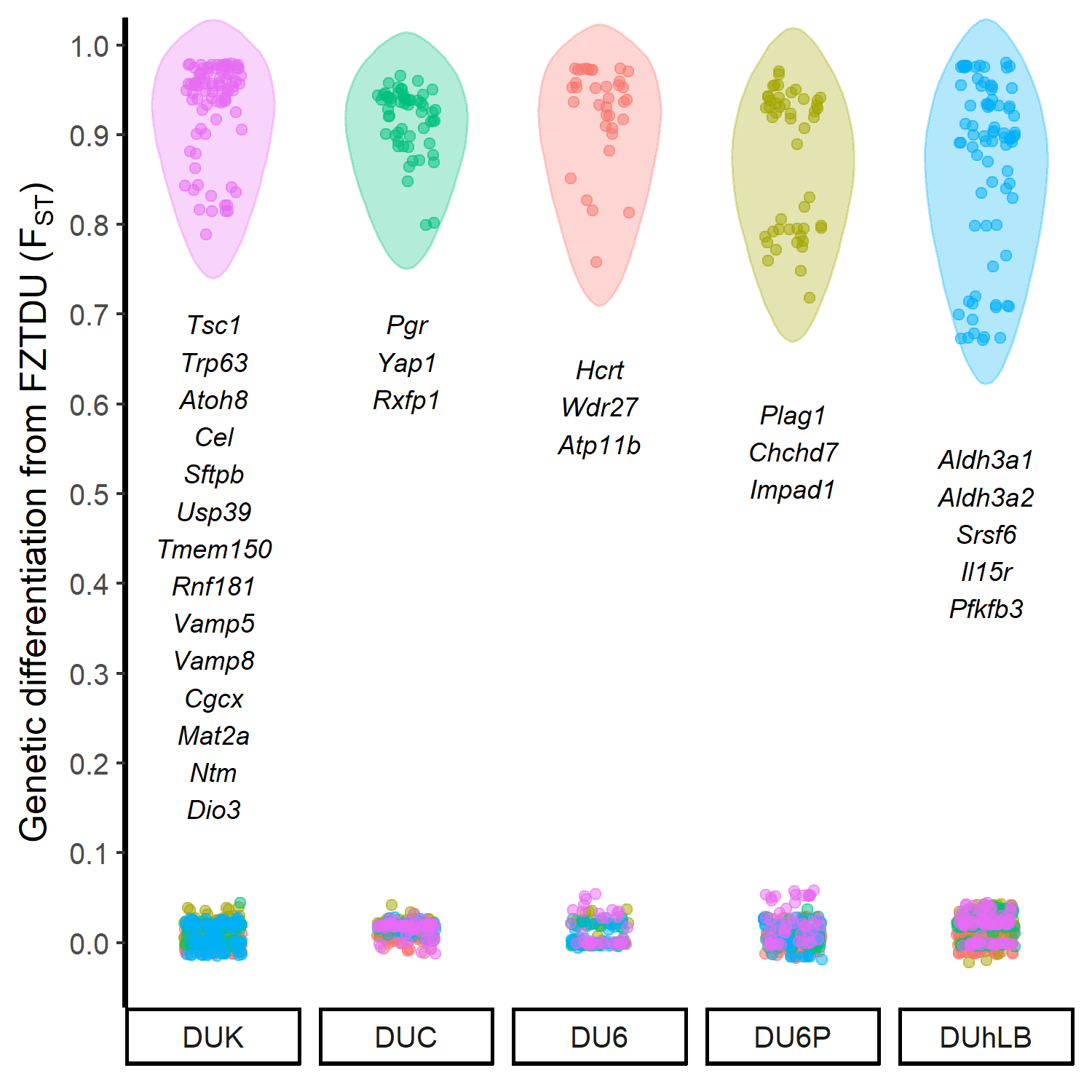
Though we understand that the lack of a proper neutral model prevents us from taking these results as final, these are promising discoveries awaiting validation. We hope that within the next years, the hidden trait-associated alleles within the genomes of these unique mouse models can be uncovered.
References
1. Palma-Vera SE, Reyer H, Langhammer M, Reinsch N, Derezanin L, Fickel J, et al. Genomic characterization of the world’s longest selection experiment in mouse reveals the complexity of polygenic traits. BMC Biol. 2022;20:52.
Follow the Topic
-
BMC Biology

This is an open access journal publishing outstanding research in all areas of biology, with a publication policy that combines selection for broad interest and importance with a commitment to serving authors well.
Related Collections
With Collections, you can get published faster and increase your visibility.
Organoids: advancements in normal development and disease modeling, and Regenerative Medicine
BMC Biology is calling for submissions to our Collection on Organoids: advancements in normal development and disease modeling, and Regenerative Medicine. This Collection seeks to bring together cutting-edge research on the use of organoids as models of normal organ development and human disease, as well as transplantable material for tissue regeneration and as a platform for drug screening.
Studies can be based on organoids derived from either induced pluripotent stem cells or tissue-derived cells (embryonic or adult stem cells or progenitor or differentiated cells from healthy or diseased tissues, such as tumors).
We welcome submissions focusing on studies investigating the mechanisms of self-organization and cellular differentiation within organoids, and how these processes recapitulate human tissue architecture and pathology. We are especially interested in studies addressing the issues of improving tissue patterning, specialization, and function, and avoiding tumorigenicity after transplantation of organoids. We will also consider studies that demonstrate the application of organoids in personalized medicine, such as drug screening, toxicity testing, and the identification of novel therapeutic targets.
We are interested in studies focusing on the refinement of methods to enhance the fidelity and functional maturity of organoids, especially those integrating organoid models with cutting-edge technologies such as advanced imaging, single-cell and spatial omics, microfluidic chip systems and bioprinting.
This Collection supports and amplifies research related to SDG 3: Good Health and Well-Being.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Mar 15, 2026
Environmental microbiology
BMC Biology is calling for submissions to our Collection on Environmental microbiology. Environmental microbiology is a rapidly evolving field that investigates the interactions between microorganisms and their surrounding environments, including plants, soil, water, and air. This area of research encompasses a diverse range of organisms, from bacteria and protists to extremophiles, and seeks to understand their roles in various ecological processes. By examining microbial communities and their functions, researchers can gain insights into plant-microbe interactions, biogeochemical cycles, nutrient cycling, and ecosystem dynamics. Furthermore, the study of the microbiome in different habitats is crucial for understanding biodiversity, ecosystem resilience, and the potential applications of microbes in environmental remediation. Advancements in molecular biology and bioinformatics have significantly enhanced our understanding of microbial ecology and the intricate relationships that underpin environmental systems. Understanding these interactions is essential for addressing pressing global issues such as climate change, pollution, and ecosystem degradation to develop sustainable strategies for environmental conservation and restoration.
Potential topics include but are not limited to:
Plant-associated microbes in sustainable agriculture
Microbiomes and symbioses in aquatic ecosystems
Microbial contributions to biogeochemical cycles
Community structure and dynamics in soil, water, air, and extreme environments
Extremophiles and their ecological significance
Pathogen Ecology
Host-Microbe Environmental Interactions
Effects of climate change and environmental stressors on microbial communities
Methodological Advances in environmental microbiology
This Collection supports and amplifies research related to SDG 6: Clean water and Sanitation, SDG 13: Climate Action, SDG 14: Life Below Water, and SDG 15: Life on Land.
All manuscripts submitted to this journal, including those submitted to collections and special issues, are assessed in line with our editorial policies and the journal’s peer review process. Reviewers and editors are required to declare competing interests and can be excluded from the peer review process if a competing interest exists.
Publishing Model: Open Access
Deadline: Apr 25, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in