TRIM5α: an unexpected journey
Published in Microbiology

The outcome of viral infections depends on a complex competition between the viruses and their hosts, and in particular the balance between host-encoded dependency factors whose exploitation promote virus growth, and immune and inflammatory responses that control infection.
The type I interferon (IFN) response is a critical element of the innate immune system that results in the transcriptional upregulation of hundreds of IFN-stimulated genes (ISGs) with diverse functions that establish a generalized antiviral state through myriad mechanisms1.
It has been previously described that IFNα pre-treatment of different cultured human cells induced a potent block to HIV-1 replication at the early stages of HIV-1 infection mainly by blocking reverse transcription and cDNA accumulation2. Despite the ubiquitous use of IFN for studying HIV-1 infection inhibition and the discovery of MX2 and other proteins as ISGs mediating cellular resistance to HIV-13, the antiviral effector mechanisms of IFN were poorly understood. Moreover, none of the described ISGs explained the IFNα-mediated block to HIV-1 reverse transcription.
When I first joined the Malim lab at King’s College London (KCL), I was interested in understanding the interplay between type I IFN and HIV-1. Therefore, the identification of host factors involved in the IFN-induced block to HIV-1 infection was the perfect project to carry out my postdoctoral training.
The first challenge I encountered with this project was finding the correct working model. We would need a cell line with two necessary features:
- Significant restriction to HIV-1 infection after IFN treatment
- Susceptibility to siRNA-mediated knockdown
After a few preliminary experiments, U87-MG cells stably expressing the CD4 and CXCR4 virus entry receptors were chosen. At this point a critical part of the work was done, as finding the best experimental model is probably the most important part when starting a new project.
Ectopic overexpression of individual ISGs was a successful approach in the past, leading to the finding of MX2 and other ISGs, but, as a gene-by-gene strategy, may miss the identification of either inhibitors that function in combination with other proteins, or positive regulators of inhibitory genes.
Therefore, to undertake a more genome-wide approach to potentially identify new ISGs that suppress HIV, we decided to use the opposite strategy. In this case, we used specific siRNAs to silence each ISG, one by one, in cells treated with IFNα. To this end, we designed a custom siRNA library targeting over 600 ISGs, and I couldn’t wait to start the experiments.
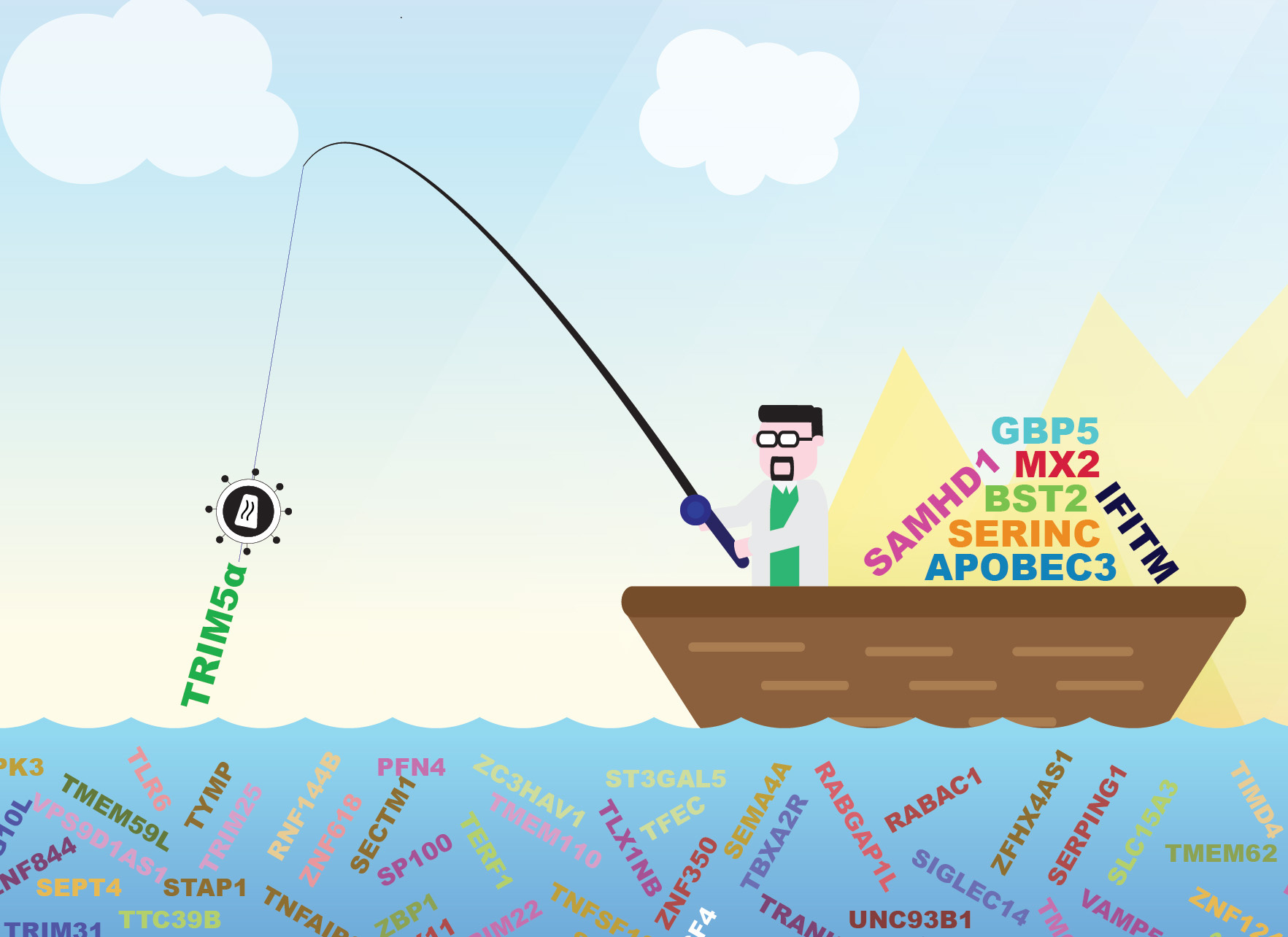
Doing a high-throughput loss-of-function screen is similar to going fishing but without the relaxing part! In a cell full of ISGs, there is no doubt that the key to a successful fishing trip is your choice of bait. Therefore, we decided to use a full-length replication-competent reporter HIV-1, to avoid any possible confounding effects that might be due to pseudotyping or the lack of accessory genes.
Using this approach, a number of genes emerged as potential candidates for further studies but, among them was human TRIM5α -- which was totally unexpected. Since the identification in 2004 of the TRIM5α protein of rhesus macaques as a restriction factor preventing HIV-1 infection in these monkeys4. Human TRIM5α, in contrast to many non-human orthologues, has not generally been ascribed significant HIV-1 inhibitory function, been regarded as virtually inactive against HIV-1.
Challenging the dogma that retroviruses have evolved to evade the TRIM5α ortholog present in species to which they are endemic required confirming our unexpected results using more than one approach. Therefore, to be confident about our findings we performed different but complementary experiments using siRNA, shRNA, CRISPR/Cas9 and ectopic overexpression in presence of IFN.
One of the hallmarks of this project was the experiment showing that TRIM5α function is operative in CD4+ T cells, the principal in vivo cell target of HIV-1 infection, invoking that TRIM5α contributes to the immune control of HIV-1 in infected humans. Working with primary cells was a difficult and arduous process due to their limited lifespan and the necessity of obtaining pure populations of CD4+ T cells expressing the indicated shRNA. However, thanks to the insertion of a fluorescent reporter and to the cell sorting facility at KCL, the experiments were successfully performed, being the most rewarding ones during this project.
Although the reliance of TRIM5α function upon IFN was a novel observation, one of the best things of working with TRIM5α is that we could take advantage of all the knowledge generated by many laboratories during the last decade, leading us to identify HIV-1 Capsid as the determinant of IFN-induced TRIM5α restriction, and to demonstrate the importance of ubiquitination.
However, the link between the described TRIM5α-mediated restriction of HIV-1 and the requirement of IFN was still missing. Although I would like to say that finding the requirement for activated immunoproteasomes for TRIM5α restriction of HIV-1 was a “eureka moment”, it was the result of hours and hours of reading very interesting manuscripts and many unsuccessful experiments examining other components of the IFN system. Together with the initial screening, finding the immunoproteasome requirement was probably the most satisfying moment during the development of this project.
Though many additional questions are posed by this paper, our work sheds significant new light not only on the biology of TRIM5α and the inhibition of HIV-1 by IFN, but also on the potential of the immunoproteasome to fine tune cellular protein function.
Jose M. Jimenez-Guardeño
The paper in Nature Microbiology is here: https://www.nature.com/articles/s41564-019-0402-0
References:
1 Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481-485 (2011).
2 Goujon, C. & Malim, M. H. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J Virol 84, 9254-9266 (2010).
3 Doyle, T., Goujon, C. & Malim, M. H. HIV-1 and interferons: who's interfering with whom? Nat Rev Microbiol 13, 403-413 (2015).
4 Stremlau, M. et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427, 848-853 (2004).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in