Tumor-educated platelets cannot be validated for breast cancer detection
Published in Cancer
The introduction of mammographic screening has enabled the detection of breast cancer in asymptomatic women, both in the general population, as well as in women with an increased risk of breast cancer who are screened from a young age onwards. Breast cancer screening has increased early detection rates, thereby improving treatment opportunities and decreasing morbidity and mortality. However, mammography is associated with a high number of false positives, leading to unnecessary invasive diagnostic procedures. In addition, the sensitivity of mammography is limited in women with high breast density, which may complicate screening of young women with an increased risk of breast cancer[1, 2].
Blood-based biomarkers have great potential, both in general cancer screening and in breast-cancer screening specifically.A variety of new technologies based on blood biomarkers are currently in development or in clinical use [3]. In recent years, tumor-educated platelets (TEPs) were considered to be an example of such a biomarker with great promise for early detection of several different cancer types. While TEP-based classifiers showed encouraging results initially [4], it is essential to perform replication studies to confirm classifier performance and robustness of the underlying methodology. Here, we assessed the value of TEPs for the detection of breast cancer in a multicenter study with an independent validation cohort, while thoroughly investigating variance related to sample origin and other potential biases within the data collected.
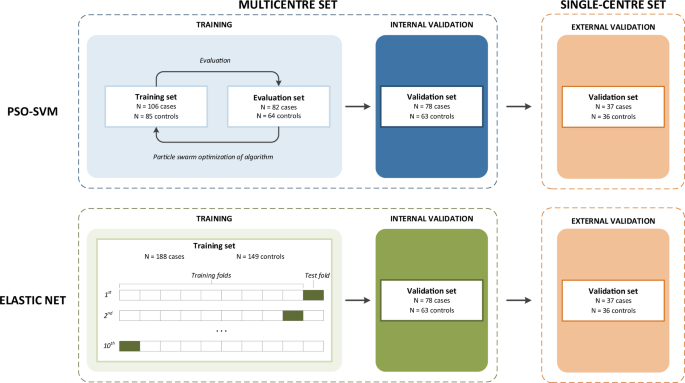
Our study focuses on the development of a classification algorithm on TEP mRNA profiles to distinguish patients with breast cancer from healthy controls. We chose to focus on a single cancer type and to avoid other non-cancer illnesses among controls to create a straightforward performance baseline, and to eliminate potential confounders within the data. In six centers, blood samples were collected and platelets were isolated according to the published protocol [5]. We included 266 women with stage I-IV breast cancer and 212 female controls. Subsequently, RNA isolation and sequencing were done centrally. Using 71% of the multicenter study population, a classifier was trained according to the published protocol by Best et al, using a particle swarm optimized support vector machine (PSO-SVM)[5]. To establish a baseline for comparison, an alternative classifier was trained using elastic net regression (EN). The performance of both classifiers was assessed using the remaining 29% of the study population. Lastly, classifier performance was validated in an independent set (37 cases and 36 controls), for which samples were collected exclusively in one of the participating centers.
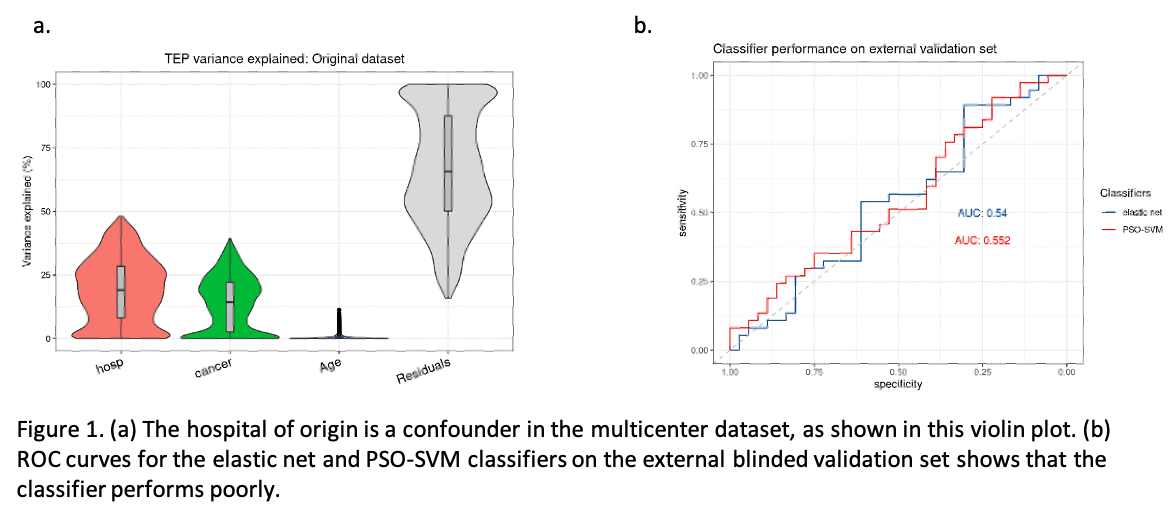
Initially, our results looked promising: both classifiers reached an area under the curve (AUC) of 0.85 upon internal validation. Post hoc analyses indicated that 19% of the variance in gene expression was associated with the hospital of origin. Genes related to platelet activity were differentially expressed between samples originating from different hospitals. Due to lack of universal sample availability, cases and controls were distributed unevenly between participating centers. A single center contributed over 68% of all controls, while 64% of cases originate from another individual center. This raised the possibility that batch effects related to hospital of origin were inflating classifier performance. We applied two different batch correction methods, Combat and RUV, to correct batch effects related to sample origin. Combat achieved a substantial reduction in variance associated with the hospital of origin, but also reduced biological signal and classifier performance. RUV batch correction preserved high classifier performance, but was unable to remove the undesired variation due to hospital of origin.
Having observed a clear batch effect related to hospital of origin that could not be corrected without loss of classifier performance, we set up a new, independent, external validation case-control study (37 cases and 36 age-matched controls) in one single center. However, both classifier types performed poorly with an AUC of 0.55 and 0.54 for the PSO-SVM and EN classifiers, respectively. Importantly, even retraining the classifier to include only samples that originated from the same center as the validation cohort did not rescue classifier performance. This suggests that centralized data cohorts that avoid hospital-related variance may still suffer from performance drop due to batch effects related to the time of sample collection.
We concluded that despite the initially good performance of the classifiers, post-hoc analyses warranted further investigation into hospital of origin as a potential confounder. Our results reveal several issues with the current TEP protocol that need to be addressed in future studies. These recommendations, such as balanced study design and standardization between labs, are important for other blood-based biomarker studies and in particular TEP based studies.
In addition to giving specific recommendations, we demonstrate the importance of publishing negative results, that highlight the challenges of molecular biomarker development. There is a tendency within academia to over-valorize and under-examine promising positive results with significant associations. However, it is equally important both for the scientific community and for patient welfare to openly publish negative results . Doing so can only promote faster development of truly revolutionary patient care while avoiding expensive and time-consuming scientific dead ends. We appreciate the opportunity the British Journal of Cancer gave us to publish this negative validation study, and would like to urge other journals to accept negative studies as well. This is particularly important with studies on novel technologies in a flourishing field like blood-based biomarkers.
References
- Myers, E.R., et al., Benefits and Harms of Breast Cancer Screening: A Systematic Review. Jama, 2015. 314(15): p. 1615-34.
- Welch, H.G., et al., Breast-Cancer Tumor Size, Overdiagnosis, and Mammography Screening Effectiveness. N Engl J Med, 2016. 375(15): p. 1438-1447.
- Ignatiadis, M., G.W. Sledge, and S.S. Jeffrey, Liquid biopsy enters the clinic — implementation issues and future challenges. Nature Reviews Clinical Oncology, 2021. 18(5): p. 297-312.
- Best, M.G., et al., RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell, 2015. 28(5): p. 666-76.
- Best, M.G., et al., RNA sequencing and swarm intelligence-enhanced classification algorithm development for blood-based disease diagnostics using spliced blood platelet RNA. Nat Protoc, 2019. 14(4): p. 1206-1234.
Follow the Topic
-
British Journal of Cancer

This journal is devoted to publishing cutting edge discovery, translational and clinical cancer research across the broad spectrum of oncology.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in