
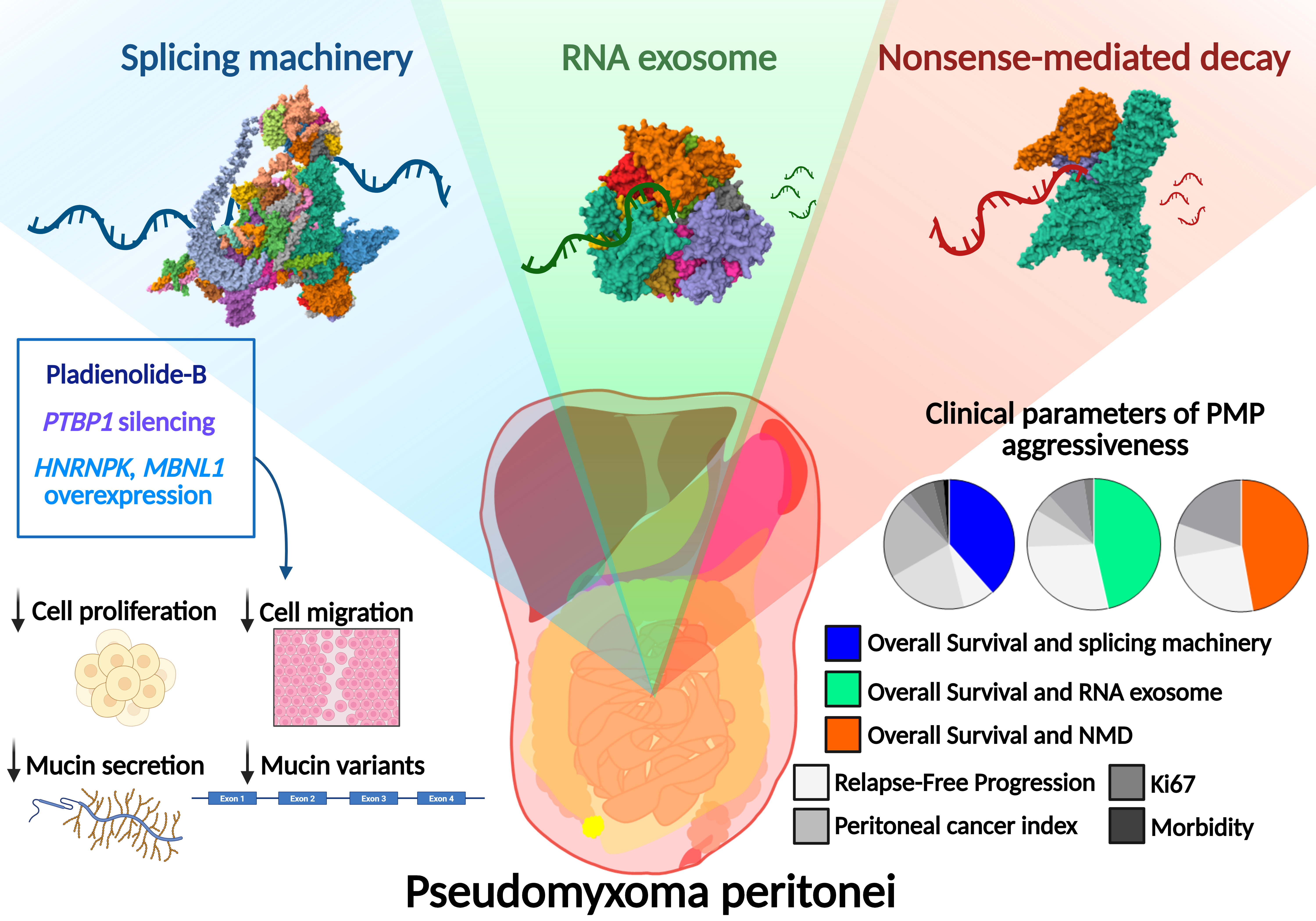
Pseudomyxoma peritonei (PMP) is a rare tumor characterized by the progressive accumulation of mucinous ascites in the abdominal cavity, most often originating from appendiceal tumors. Although it typically follows a slow-growing course, PMP remains clinically challenging due to its high recurrence rate. One of the major obstacles to understanding this disease is the mucin-rich and sparsely cellular nature of the tumor tissue, which limits molecular characterization and has historically hindered research efforts.
While previous studies have focused on signaling pathways such as TGF-β, epithelial-mesenchymal transition, and extracellular matrix remodeling, the role of RNA regulatory mechanisms in PMP has remained largely unexplored. In our study, we present the first comprehensive analysis of key RNA-related processes in PMP, including alternative splicing, the RNA exosome complex, and nonsense-mediated decay (NMD), three fundamental machineries for the regulation of the diversity, stability, and turnover of RNA molecules. Using a targeted microfluidic approach on 29 patient samples, supported by external transcriptomic and proteomic datasets, we uncovered a widespread disruption of these RNA regulatory machineries in PMP. Many of these alterations correlated with clinical parameters and allowed to distinguish tumor from normal tissue. This is the first time that such RNA dysregulation has been reported in this disease, offering new molecular insights into its pathogenesis.
To understand whether these changes had functional consequences, we conducted in vitro experiments targeting the splicing machinery. Using Pladienolide-B, a well-known pharmacological inhibitor of splicing, we found a marked reduction in tumor cell viability and migration. Remarkably, when combined with standard chemotherapeutic agents, the effects were even stronger, suggesting a potential synergistic effect. We then decided to silence or overexpress specific splicing factors to check if precise modulation of splicing machinery could reduce PMP cells aggressiveness. Silencing PTBP1, a splicing factor upregulated in PMP, reduced cell viability. Conversely, restoring the expression of HNRNPK and MBNL1, both found to be downregulated in tumors, had similar anti-tumor effects. Interestingly, splicing inhibition and modulation also influenced the expression and splicing of MUC2, the gene encoding the main mucin in PMP.
Beyond splicing, we identified alterations in two other key RNA surveillance processes: RNA exosome and NMD, which eliminates faulty transcripts that could otherwise produce aberrant proteins. These RNA-processing alterations point to a novel layer of molecular vulnerability in PMP. In many cases, the dysregulation of splicing, RNA exosome and NMD components was associated with clinical parameters of PMP aggressiveness and progression and also with the expression of other cancer-related genes.
Taken together, our results reveal, for the first time, a widespread disruption of RNA-regulatory pathways in PMP linked to disease progression and tumor aggressiveness. Splicing inhibition or modulation of specific splicing factors, emerges as a promising strategy to reduce aggressive tumor behavior and enhance the effect of existing therapies. These findings open new avenues for exploring RNA biology as a source of clinically actionable targets in this rare and understudied cancer.
Follow the Topic
-
Cancer Gene Therapy

The essential gene and cellular therapy resource for cancer researchers and clinicians, keeping readers up to date with the latest developments in gene and cellular therapies for cancer.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in