Understanding the impact of Enteromyxum leei on gilthead seabream and its microbiota
Published in Ecology & Evolution, Microbiology, and Protocols & Methods

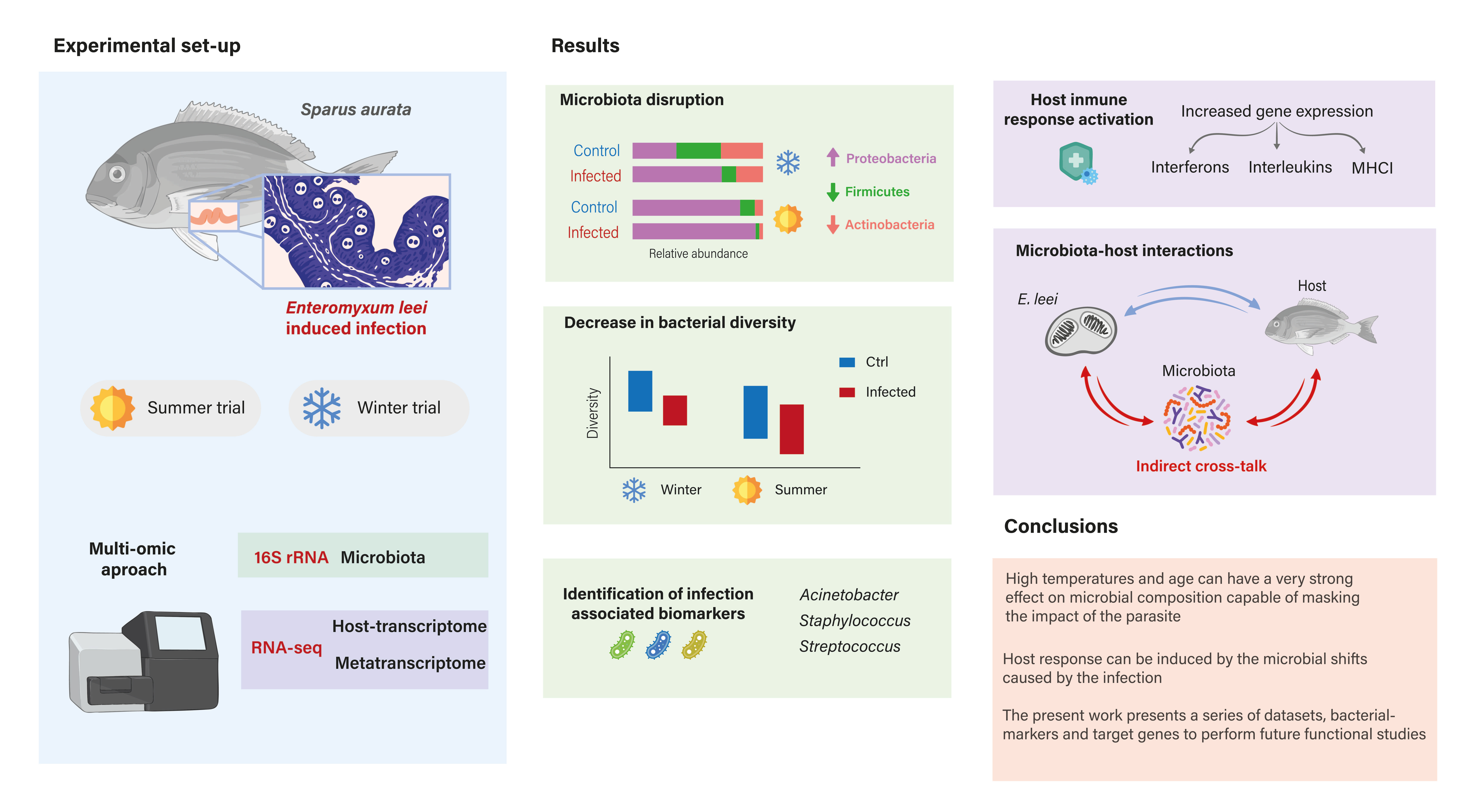
Background Gilthead seabream (Sparus aurata) is a major fish species for Mediterranean aquaculture, but its production faces challenges due to diseases. One of them is enteromyxosis, a disease caused by the intestinal parasite Enteromyxum leei. It leads to severe economic losses by reducing growth rates, triggering intestinal inflammation, and increasing mortality rates. Currently, no effective treatments exist for enteromyxosis, which makes it crucial to understand its effects on fish health and microbiota.
Study Objectives In this study we explored the interactions between the fish host, its intestinal microbiota, and E. leei infection using a multi-omic approach. This means that we integrated metataxonomics (study of microbial communities), host transcriptomics (gene expression analysis), and metatranscriptomics (gene activity in microbiota) to assess changes caused by infection.
Key Findings
- Microbiota Disruption: Infection with leei significantly altered the gut microbiota of gilthead seabream. Infected fish, during the winter season, exhibited an increase in harmful bacteria such as Proteobacteria (rising from 32.3% in healthy fish to 89.8% in infected fish). Conversely, beneficial bacteria like Firmicutes and Actinobacteria decreased sharply.
- Microbial Biomarkers Identified: The genus Acinetobacter emerged as a key indicator of infection, while beneficial bacteria like Streptococcus and Staphylococcus, which may contribute to gut health, were significantly reduced in infected fish. Identifying these microbial markers could help develop diagnostic tools for early detection of enteromyxosis.
- Host Immune Response Activation: Infected fish exhibited increased expression of 935 genes in the intestine, many related to immune system functions, including pathways related with interferons, interleukins, and MHCI, which play a role in fighting infections. The spleen, an organ involved in immunity in fish, however, showed minimal changes.
- Temperature and Age Influence: We found that seasonality and fish age also played a role in modulating microbial composition. During the summer season, even uninfected fish displayed altered gut microbiota, masking some parasite-related changes. Younger fish appeared to have a more resilient microbiota, suggesting that age-related factors might influence disease susceptibility and gut microbial diversity.
- Microbiota-Host Interactions: Some immune responses in infected fish were not directly caused by leei, but were instead linked to changes in the microbiota. The decrease in beneficial bacteria, which help regulate inflammation, may have contributed to immune activation and inflammation. This finding underscores the importance of maintaining a balanced microbiota to support fish health.
- Potential Strategies for Disease Control: Given the absence of effective treatments, the study suggests that future disease management should focus on preventative approaches. Probiotics, dietary modifications, and environmental management (e.g., temperature control) could be employed to enhance microbial diversity and boost natural immunity against leei infections.
- Conclusions and Implications Our study highlights the complex relationship between the gut microbiota, host immune system, and parasite infections in aquaculture. The findings suggest that monitoring gut microbiota composition could serve as an early indicator of fish health issues. Additionally, strategies to support beneficial gut bacteria—such as probiotics or dietary modifications—could improve fish resilience against leei and other pathogens. By understanding these interactions, aquaculture stakeholders can develop better disease management strategies, ultimately leading to healthier fish and more sustainable production practices. The integration of microbiome-based strategies into aquaculture health management could pave the way for innovative, non-invasive solutions that mitigate the economic and ecological impact of enteromyxosis.
Follow the Topic
-
Animal Microbiome

This is a community-focused journal welcoming all animal microbiome studies relating to domestic and non-domestic animals. The journal welcomes submissions which go beyond measurements of diversity and move towards a functional understanding of animal-associated microbiomes.
Related Collections
With Collections, you can get published faster and increase your visibility.
Animal Gut Nutrition and Greenhouse Gas Mitigation
Animal Microbiome and Journal of Animal Science and Biotechnology call for submissions to the collection on Animal Gut Nutrition and Greenhouse Gas Mitigation.
Efforts to reduce greenhouse gas emissions from livestock systems increasingly hinge on innovations in animal gut nutrition. The dynamic relationship between the gut microbiome and nutrient utilization plays a pivotal role in shaping methane output, feed efficiency, and overall sustainability. Advances in microbial ecology—particularly in understanding the role of gut microbiome in nutrient metabolism—are opening new pathways for mitigating emissions while enhancing productivity. These developments support the implementation of climate-smart agricultural strategies to address climate change and its impacts.
Looking ahead, continued research in this field has the potential to yield innovative solutions such as targeted probiotic supplementation, which could further optimize gut function and enhance nutrient absorption. These advancements may lead to reduced greenhouse gas emissions while improving animal health and productivity. By deepening our understanding of the animal gut microbiome, we can contribute significantly to sustainable agricultural practices that benefit both the environment and food security.
We invite researchers to contribute to this special Collection on Animal Gut Nutrition and Greenhouse Gas Mitigation. Topics of interest include but are not limited to:
- Animal Gut Microbiome and Feed Efficiency
- Greenhouse Gas Mitigation Strategies
- Rumen Fermentation Dynamics
- Nutrient Utilization in Livestock
- Probiotic Supplementation Effects
- Sustainable Livestock Production Practices
- Climate-Smart Agriculture Innovations
This Collection supports and amplifies research related to SDG 13, Climate action.
All submissions in this collection undergo the relevant journal’s standard peer review process. Similarly, all manuscripts authored by a Guest Editor(s) will be handled by the Editor-in-Chief of the relevant journal. As an open access publication, participating journals levy an article processing fee (Animal Microbiome fees, Journal of Animal Science and Biotechnology fees). We recognize that many key stakeholders may not have access to such resources and are committed to supporting participation in this issue wherever resources are a barrier. For more information about what support may be available, please visit OA funding and support, or email OAfundingpolicy@springernature.com or the Editor-in-Chief of the journal where the article is being submitted.
Publishing Model: Open Access
Deadline: Sep 04, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in