Unearthing the Resilience: How Desert Rhizospheres Tell Two Microbial Tales
Published in Ecology & Evolution and Microbiology
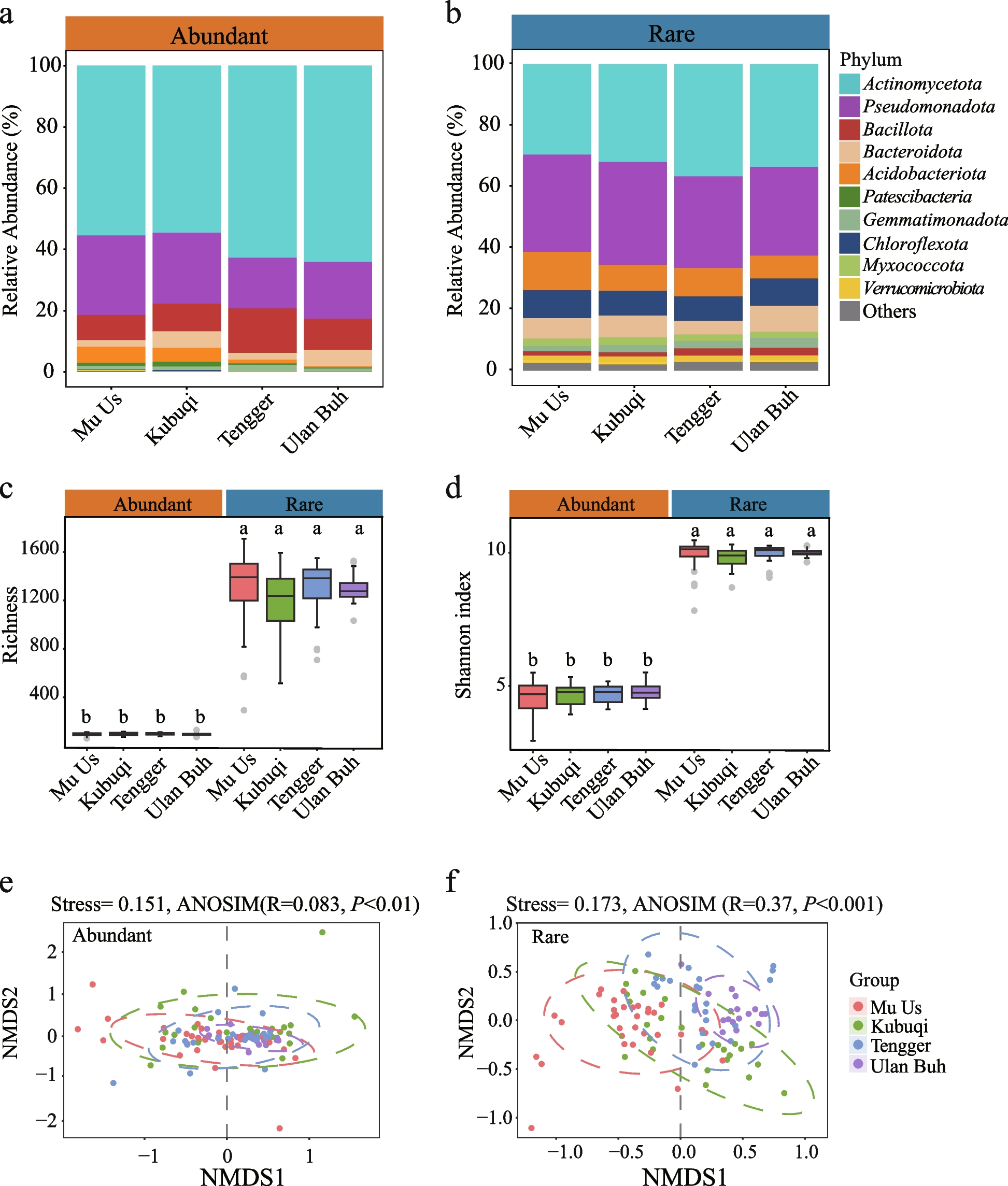
Our research team has long focused on desert ecosystems, not only for their stark and majestic beauty, but also because they represent one of the most extreme and underexplored frontiers in microbial ecology. During our 2022 field investigations in sites dominated by Artemisia desertorum, a drought-adapted desert shrub, we observed a compelling phenomenon: while the surface soils appeared almost lifeless, the rhizosphere zones harbored surprisingly rich and active microbial communities. This led us to a deceptively simple question: How do rare and abundant microbial taxa within the rhizosphere respond differently to drought stress? At the time, rare taxa were often dismissed as ecological ‘background noise,’ but we viewed them as potentially significant ecological contributors. The true challenge of this research lay not only in enduring 45°C sampling conditions—though that was undoubtedly taxing—but in extracting meaningful ecological signals from layers of technical and biological complexity.
Rhizosphere microbial communities are inherently intricate, and defining what constitutes “rare” versus “abundant” taxa, standardizing the data, and determining robust abundance thresholds sparked intense discussion within our team. A pivotal shift in our study occurred when we stopped treating ‘rare’ and ‘abundant’ solely as statistical labels and began considering them as ecologically distinct groups with potentially divergent functions. By integrating amplicon sequencing data with environmental gradients and climatic variables, we found that rare taxa, though low in abundance, were more sensitive to drought and exhibited stronger environmental filtering. In contrast, abundant taxa appeared more stable and potentially more functionally redundant. These findings prompted us to rethink conventional views of the ‘core microbiome’ and recognize the nuanced resilience strategies embedded within plant–microbe interactions in extreme environments. Perhaps the most difficult part of writing the manuscript was not a lack of findings, but rather deciding what to focus on. We uncovered many intriguing patterns, but ultimately chose to center the narrative on the divergent drought responses of rare versus abundant taxa—not only because it was novel, but because it was ecologically meaningful.

As climate extremes become more frequent, understanding microbial community structure alone is insufficient—we must also ask: Who is responding? How are they responding? And why? This study reinforced a central ecological insight: even the seemingly inconspicuous “minor players” in microbial communities can hold critical roles in ecosystem stability. In the quiet microzones of the desert rhizosphere, resilience unfolds in ways we are only beginning to understand.
Follow the Topic
-
Advanced Biotechnology

This journal is open access journal committed to publish cutting-edge research in the field of biotechnology. It aims to provide a platform to advance knowledge, techniques and strategies in biotechnology by covering all the relevant fields of biotechnology.
Related Collections
With Collections, you can get published faster and increase your visibility.
Artificial Intelligence in Life Sciences
Artificial intelligence (AI) technologies are profoundly transforming research paradigms in life sciences, providing innovative solutions for analyzing complex biological systems, uncovering disease mechanisms, and advancing precision medicine. In recent years, AI methodologies, especially deep learning, have achieved groundbreaking progress in genomics, proteomics, drug discovery, and medical imaging.
This special issue aims to systematically synthesize AI-driven technologies in life sciences, establish interdisciplinary methodological frameworks, accelerate data-to-knowledge translation, and promote cross-disciplinary integration in AI-powered life science research. We invite submissions of all article types, including original research papers, reviews, protocol, letter, methodology, and case study, with emphasis on: AI algorithm development, multimodal data integration (e.g., multi-omics, imaging, and clinical records), interpretable model construction, and biomedical implementation.
The scope of this issue includes, but is not limited to:
1. AI for biological big data mining
2. Biomolecular structure prediction and design
3. Intelligent breeding and gene editing optimization
4. AI-accelerated drug discovery
5. Precision medicine and disease prediction
6. Synthetic biology and biosystem modeling
7. Biomedical knowledge graph construction
8. Explainable AI (XAI) and biological model reliability
9. AI analysis of single-cell and spatial omics technologies.
If you have interest to submit a paper to this special issue, or have any question about this topic, please do not hesitate to contact the journal editor (advbiotech@mail.sysu.edu.cn).
Publishing Model: Open Access
Deadline: Sep 30, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in