Unlocking secrets of polyketide synthase assembly lines
Published in Chemistry

Modular polyketide synthases (PKSs) were discovered in the 1990s in bacteria and thousands of these proteins have been identified since then. The polyketide products are bioactive compounds, and include clinically important drugs such as erythromycin (antibiotic), rapamycin (immunosuppressant), and epothilone (anticancer drug) 2,3.
While the polyketides are extraordinarily diverse, they are produced by evolutionarily related proteins that share the molecular logic for polyketide biosynthesis 4. A plethora of studies performed during almost three decades could disclose many key aspects of modular PKSs: (i) Synthesis is catalyzed by a set of domains, of which C-C bond formation via Claisen condensation mediated by the ketoacyl synthase (KS) domain represents the catalytic key step, (ii) domains join together to form modules with some domains being mandatory and others optional, (iii) a carrier domain present within every module covalently binds substrates and intermediates, and (iv) most modular PKSs known to date are comprised of six or more modules and assemble to complexes of several megadaltons (Figure 1).
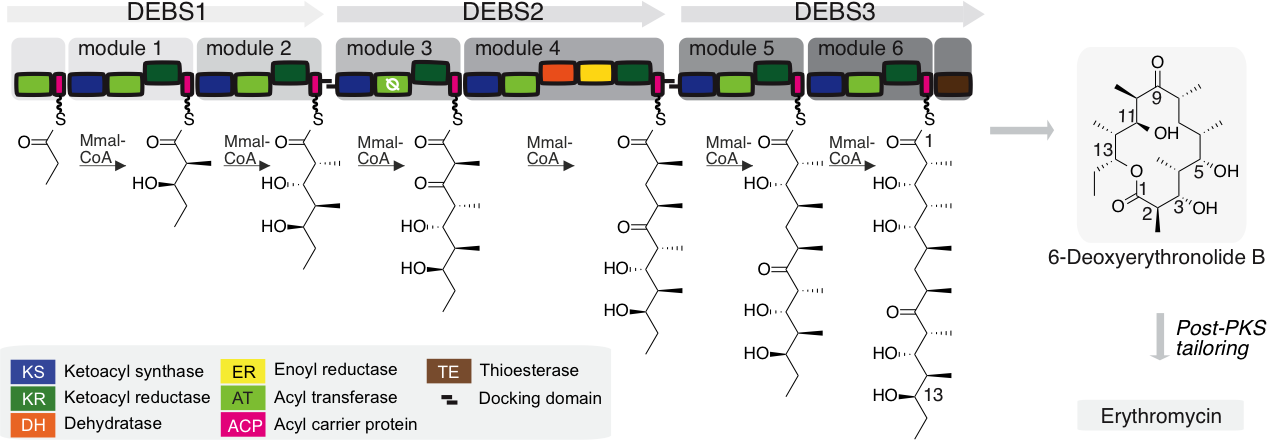
Figure 1. Assembly line synthesis by modular PKSs. Biosynthesis of 6-deoxyerythronolide B by the modular PKS 6-deoxyerythronolide B synthase. A propionyl moiety is successively elongated with methylmalonyl-CoA (MMal-CoA) by modules M1 to M6, provided on three polypeptides (DEBS1-3). Each module harbors a region for the elongation of the polyketide intermediate and can possess an additional region responsible for further processing of the elongated polyketide. 6-Deoxyerythronolide B is cyclized upon hydrolytic release. Boxes in increasing grey scale highlight consecutive modules. Catalytic domains are represented by different colors, as indicated. Docking domains enable non-covalent interactions between the polypeptides DEBS1-3.
Despite the solid understanding of modular PKSs, the mechanistic basis of directional (or vectorial) synthesis along the linearly arranged modules remains elusive. Recent structural work, in combination with functional studies, achieved new insight into how these proteins enable this synthetic feat 5,6. Independently of the type module, that means the number and nature of domains provided by the module as well as the polyketide intermediate that it processes, the critical step is the translocation of the growing polyketide to the KS domain of the downstream module for C-C bond formation and the simultaneous suppression of its re-loading into the KS of the same module (Figure 2). Intuitively, one would account substrate specificity of the condensing enzyme responsible for translocation, e.g., by the condensing enzyme of the same module preventing the elongated polyketide from re-binding, but several studies demonstrated that the substrate specificity is rather weak and cannot meet this demand (e.g. ref 7). Thus, something else must be responsible for the programming – likely a more general property that is an inherent feature of every module or emerges from the assembly of modules.
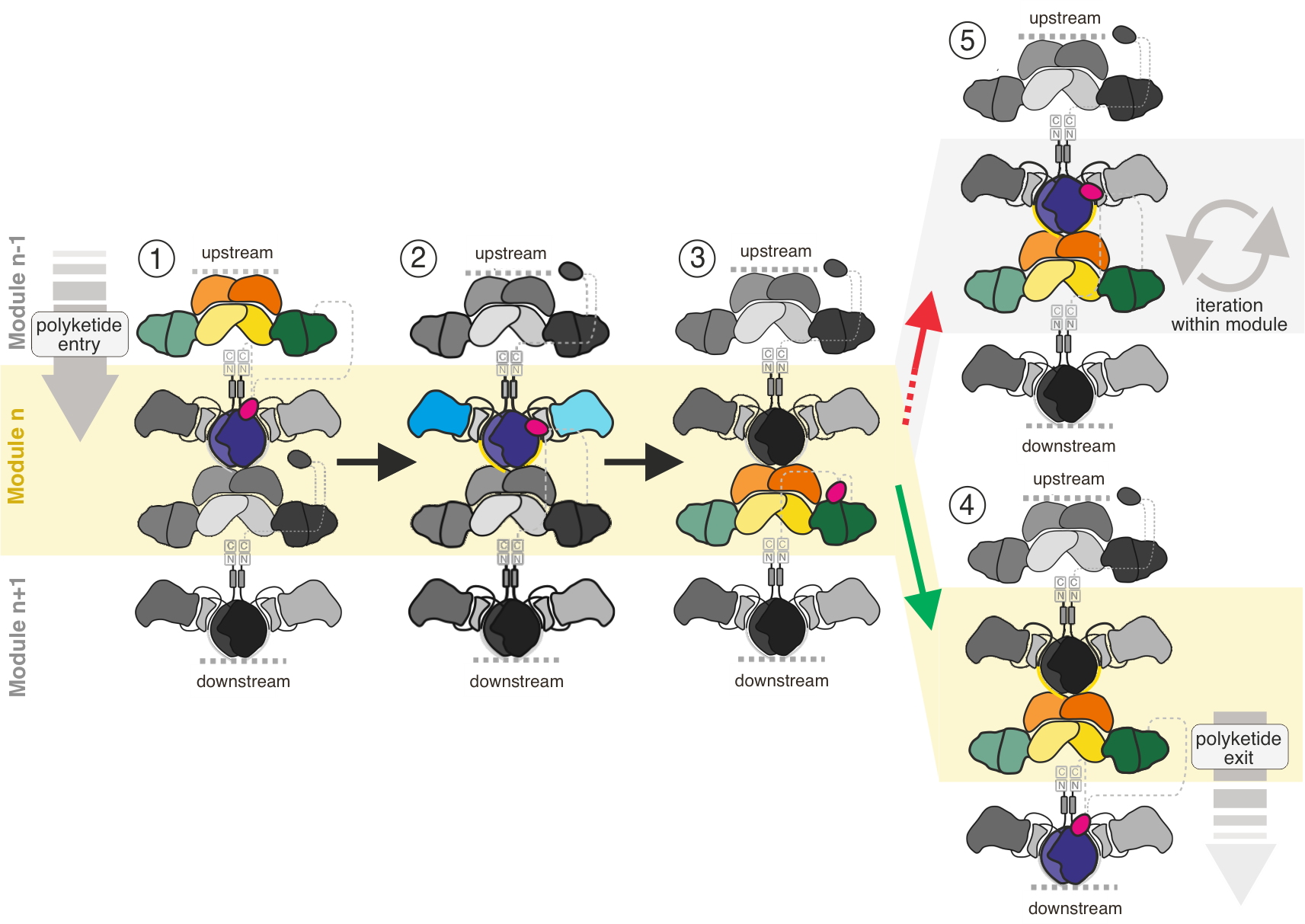
Figure 2. Vectorial vs. iterative synthesis. Polyketide processing in modular PKSs is illustrated for one module (module n, yellow background) embedded in a modular assembly line. Synthesis is shown for one half of the modules only, and complexity of the synthesis is reduced to key steps, that are: (1) entry of the polyketide from upstream, (2) KS-mediated elongation of the polyketide by two carbons, (3) processing of the elongated polyketide shown for reduction by KR only, (4) exit of the elongated and processed polyketide to the downstream module by translocation. (5) Re-loading the polyketide into the KS of the same module needs to be suppressed. Domain coloring as in Figure 1.
In order to understand the capability of each module for directed synthesis, it should be mentioned at this point that also mono-modular systems with repetitive synthesis do exist in nature. These systems comprise one set of catalytic domains, which they use for several cycles of synthesis. The most prominent representative is the fatty acid synthase (FAS), which produces fatty acids of distinct lengths by repeated elongation of acyl chains 8. Further, mostly fungi but also bacteria harbor a plethora of iterative PKSs, which like FASs perform several elongation steps within a single module, but are able to produce spectacular synthetic output by varying synthesis per cycle 9. FASs and iterative PKSs are evolutionarily related and supposed to be precursors of modular PKSs 10,11. Accordingly, the challenge is also to understand how a module at some point during PKS evolution acquired the ability to synthesize vectorially rather than iteratively.
Recent work demonstrates that the key to vectorial synthesis is the positional variability of folds, domains, and subregions of these proteins. While the high conformational dynamics of these types of proteins were revealed many years ago (for a recent work, see 12), new studies suggest that vectorial synthesis emerges from distinct and synchronized conformational changes. Two solutions for vectorial synthesis are presented: one from the labs of Fromme and Kim, based on structural and functional analysis of the lasalocid A synthase polypeptide 14 (Lsd14), and a second one by the labs of Chiu and Khosla, based on 6-deoxyerythronolide B synthase module 1 (DEBS M1).
While agreeing overall in the concept that synchronized conformations dictate vectorial synthesis, the two models differ fundamentally in their specific solution of how the translocation of the polyketide to the KS domain of the downstream module is enabled and the re-loading into the KS of the own module is prevented: (i) The DEBS M1 model suggests that the access to the own KS is blocked by the flexing of the adjacent acyl transferase (AT) domain. Flexing is fueled by the exergonic Claisen condensation and then re-established in a timely coordinated fashion with translocation. (ii) In the Lsd14 model, translocation is a consequence of positional variability of the functional domains; specifically, of the proximity of the carrier domains to different catalytic domains, thereby controlling the chances for productive interaction. Unlike DEBS M1, vectorial synthesis does not arise from a specialist structural feature but from the kinetic coupling of catalytic steps along lined-up modules.
Read more about the current understanding of these fascinating proteins at: https://www.nature.com/articles/s41589-023-01277-7 (ref 13)
Acknowledgement: Thanks to Lynn Buyachuihan for discussions and editing.
References:
- Deguchi, T. et al. Direct observation of motor protein stepping in living cells using MINFLUX. Science 379, 1010–1015 (2023).
- Staunton, J. & Weissman, K. J. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18, 380–416 (2001).
- Hertweck, C. The Biosynthetic Logic of Polyketide Diversity. Angew. Chem. Int. Ed. 48, 4688–4716 (2009).
- Nivina, A., Yuet, K. P., Hsu, J. & Khosla, C. Evolution and Diversity of Assembly-Line Polyketide Synthases: Focus Review. Chem. Rev. 119, 12524–12547 (2019).
- Bagde, S. R., Mathews, I. I., Fromme, J. C. & Kim, C.-Y. Modular polyketide synthase contains two reaction chambers that operate asynchronously. Science 374, 723–729 (2021).
- Cogan, D. P. et al. Mapping the catalytic conformations of an assembly-line polyketide synthase module. Science 374, 729–734 (2021).
- Menzella, H. G. et al. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat. Biotechnol. 23, 1171–1176 (2005).
- Heil, C. S., Wehrheim, S. S., Paithankar, K. S. & Grininger, M. Fatty Acid Biosynthesis: Chain-Length Regulation and Control. Chembiochem 20, 2298–2321 (2019).
- Cox, R. J. Curiouser and curiouser: progress in understanding the programming of iterative highly-reducing polyketide synthases. Nat. Prod. Rep. 40, 9–27 (2023).
- Wang, B., Guo, F., Huang, C. & Zhao, H. Unraveling the iterative type I polyketide synthases hidden in Streptomyces. Proc. Natl. Acad. Sci. USA 117, 8449–8454 (2020).
- Grininger, M. The role of the iterative modules in polyketide synthase evolution. Proc. Natl. Acad. Sci. USA 117, 8680–8682 (2020).
- Klaus, M. et al. Solution Structure and Conformational Flexibility of a Polyketide Synthase Module. JACS Au jacsau.1c00043 (2021) doi:10.1021/jacsau.1c00043.
- Grininger, M. Enzymology of assembly line synthesis by modular polyketide synthases. Nat. Chem. Biol. (2023) doi:10.1038/s41589-023-01277-7.
Follow the Topic
-
Nature Chemical Biology

An international monthly journal that provides a high-visibility forum for the chemical biology community, combining the scientific ideas and approaches of chemistry, biology and allied disciplines to understand and manipulate biological systems with molecular precision.

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in