Unraveling the Complexity of Gene-Environment Interactions in Noncommunicable Diseases through Multi-Omics Research
Published in Protocols & Methods, Computational Sciences, and Genetics & Genomics
By Dr. Robel Alemu (Lead Author), Associate Professor Azmeraw Amare (Senior Author), Associate Professor Tesfaye Mersha (Senior Author) and Collaborators
Understanding the Genetic and Environmental Complexity of Chronic Diseases
Noncommunicable diseases (NCDs), such as cardiovascular diseases, diabetes, cancers, and chronic respiratory conditions, are the leading causes of mortality worldwide, accounting for over 74% of global deaths
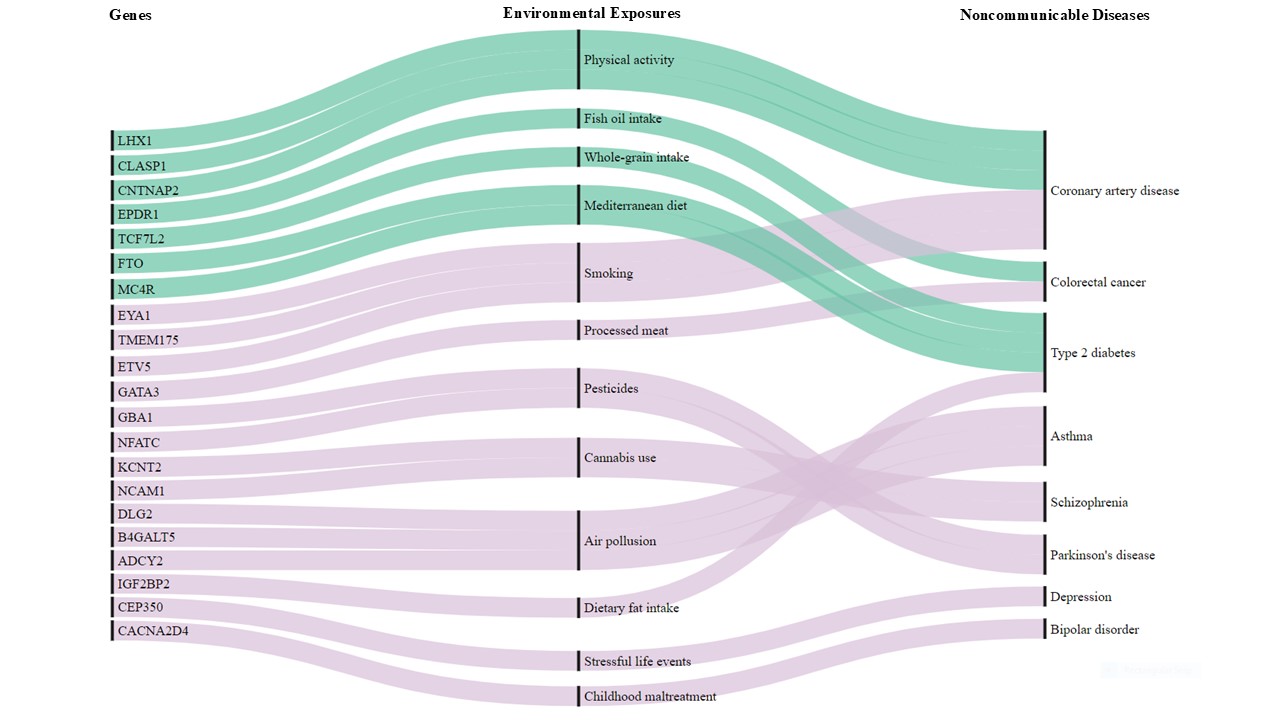
This intricate biological interplay means that individuals carrying the same genetic variant may experience different disease outcomes depending on their environment. For instance, certain genetic variants increase the risk of Parkinson’s disease in individuals exposed to pesticides
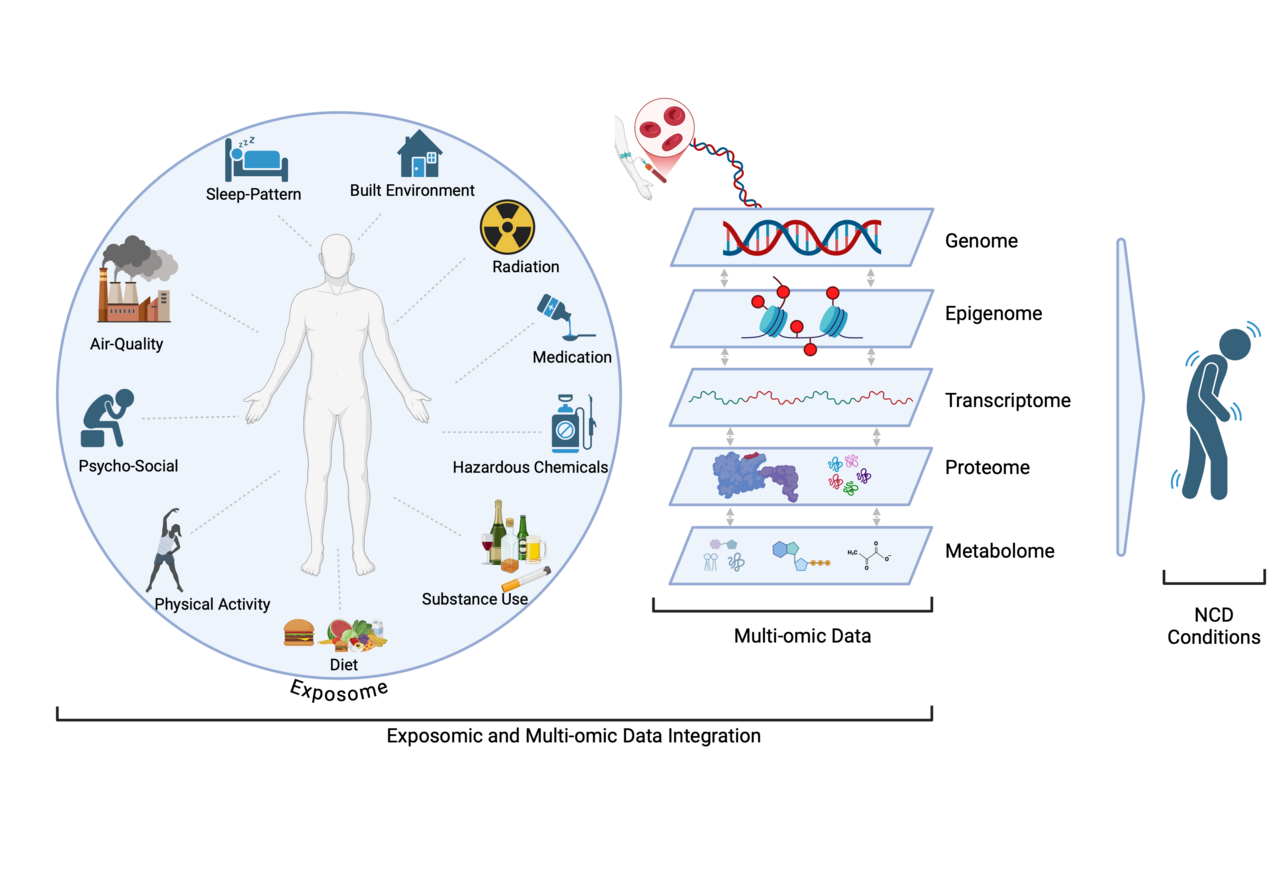
How Multi-Omics is Transforming Our Understanding of Disease Mechanisms
Genomic research, particularly genome-wide association studies (GWAS), has identified thousands of genetic variants linked to NCDs
By integrating multi-omics data with exposome data—which includes lifestyle, environmental, and social determinants of health—researchers can map how environmental influences shape biological pathways that drive disease
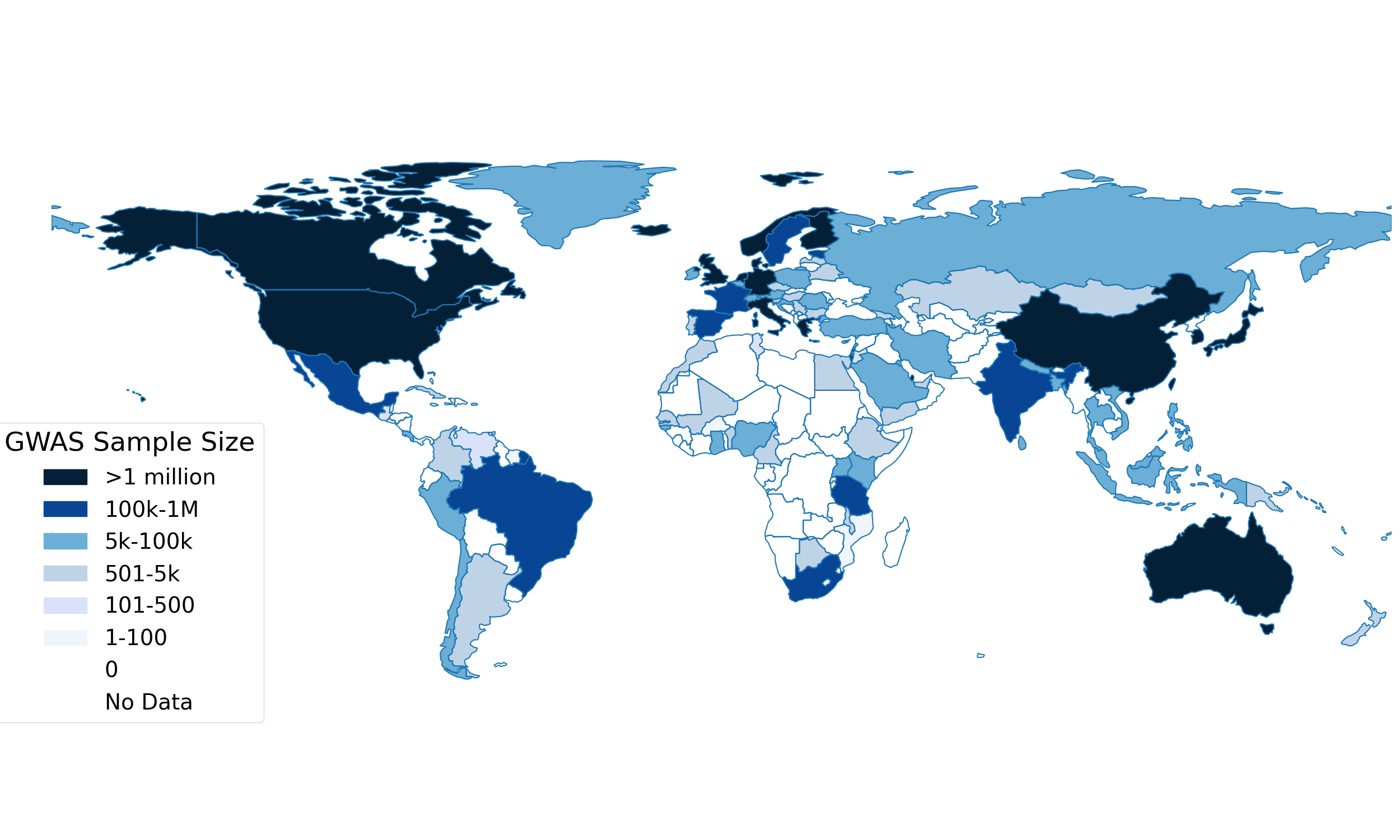
Expanding Global Representation in Omics Research
One of the most pressing challenges in genetic and multi-omics research is the underrepresentation of non-European populations. More than 85% of GWAS participants are of European ancestry
However, this diversity gap is not limited to genomic data—the disparities are even more pronounced in other omic layers, such as epigenomics, proteomics, and metabolomics
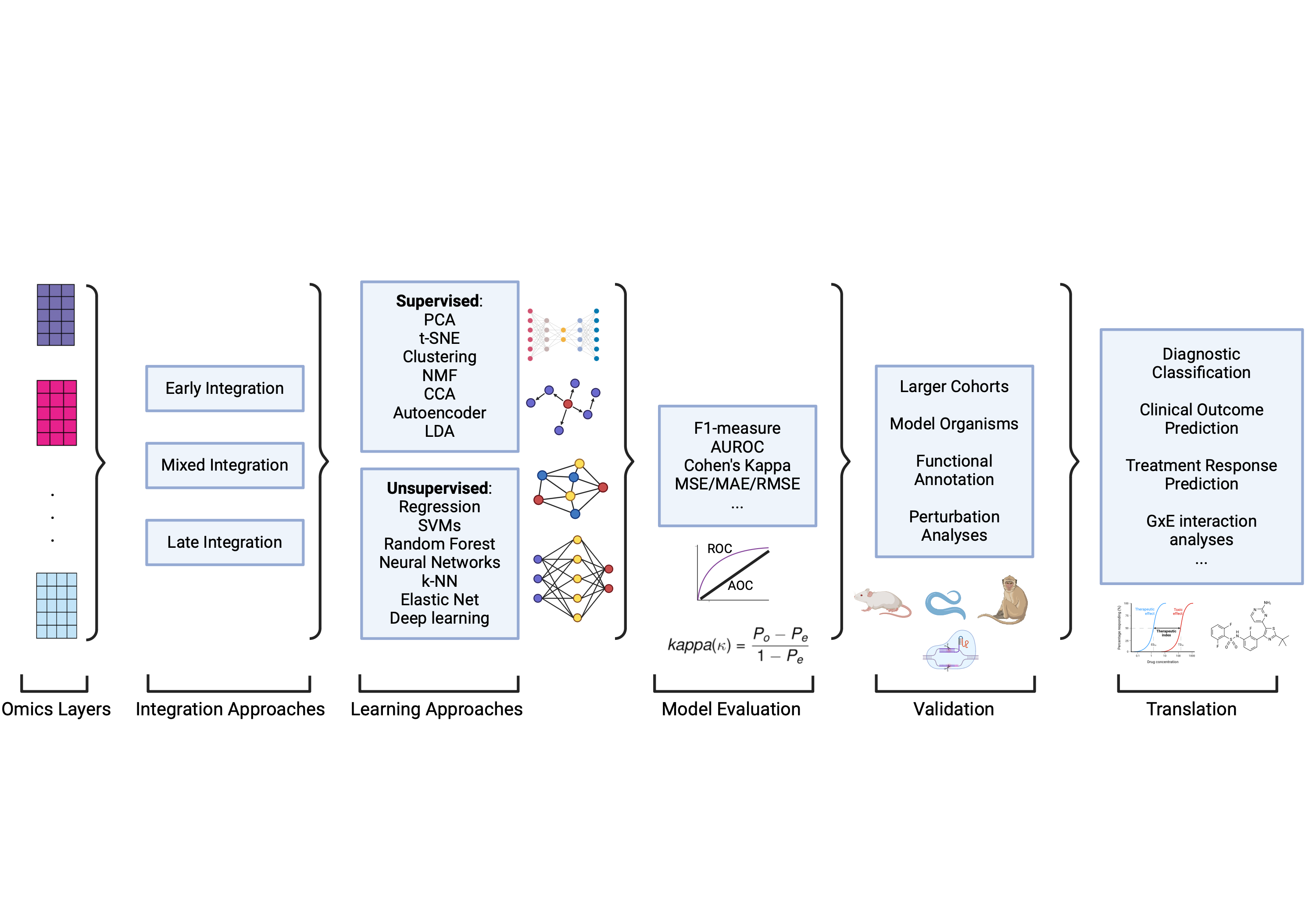
AI and Machine Learning in Multi-Omics Research: Addressing GxE Challenges
Gene-environment (GxE) interaction studies face persistent challenges, particularly limited sample sizes and the burden of multiple testing when analyzing high-dimensional biological data
AI and ML are transforming multi-omics research by enabling the integration of large, complex datasets and identifying hidden patterns that traditional methods often miss. Various computational techniques—including unsupervised learning methods like Principal Component Analysis (PCA) and t-SNE, as well as supervised approaches such as Support Vector Machines (SVMs), Random Forests, and deep learning—help uncover key biological insights by linking genetic and environmental factors to disease phenotypes
For example, deep learning models such as those developed by Wu et al. (2023) simultaneously estimate main effects and GxE interactions, overcoming hierarchical constraints in conventional regression-based models
However, AI-driven approaches are not without challenges. Bias in training datasets remains a major concern, as underrepresentation of certain populations can lead to skewed model performance and exacerbate disparities in GxE research. Additionally, the “black box” nature of deep learning models makes it difficult to interpret findings, raising concerns about clinical transparency and trust. Ethical considerations, such as data privacy and responsible AI implementation, must also be addressed to ensure equitable, reliable, and scalable applications of AI in multi-omics research.
Real-World Applications of Multi-Omics Research in Medicine
Multi-omics approaches are already shaping clinical decision-making and treatment strategies in various ways. For example, recent studies have identified genes that protect neurons from oxidative stress, a major contributor to neurodegenerative diseases like Alzheimer’s and Parkinson’s. In pharmacogenomics, multi-omics research has enabled personalized drug dosing, such as tailoring warfarin prescriptions based on CYP2C9 and VKORC1 genetic variants
These advances underscore the potential of multi-omics research to revolutionize medicine, moving us closer to individualized prevention and treatment strategies that consider both genetic makeup and environmental influences.
A Global Call to Action
To fully realize the potential of multi-omics research, scientists, policymakers, and funding agencies must prioritize global inclusivity and data-sharing initiatives. Expanding research in underrepresented populations, strengthening research infrastructure in low- and middle-income countries, improving analytical techniques for dissecting GxE interactions, and establishing global standards for data sharing and integration will be key to accelerating scientific discoveries and improving health outcomes worldwide.
As lead author Dr. Robel Alemu emphasizes, “Our review highlights the transformative power of multi-omics research in revealing the biological mechanisms behind chronic diseases. However, this potential will remain unrealized unless we address the significant equity gaps in omics research and ensure that these advancements benefit all populations.”
Join the conversation! How do you see multi-omics shaping the future of precision medicine? Let us know your thoughts in the comments! 🚀
Robel Alemu (Ph.D.)
Postdoctoral Researcher, University of California Los Angeles
Broad Institute of MIT and Harvard; The University of Adelaide Medical School
Email: robel.alemu@anderson.ucla.edu; ralemu@broadinstitute.org
References
-
- World Health Organization. Global Action Plan for the Prevention and Control of Noncommunicable Diseases, 2013-2020.
- Calcaterra, V. & Zuccotti, G. Non-Communicable Diseases and Rare Diseases: A Current and Future Public Health Challenge within Pediatrics. Children vol. 9 Preprint at https://doi.org/10.3390/children9101491 (2022).
- Ngo, K. J. et al. Lysosomal genes contribute to Parkinson’s disease near agriculture with high intensity pesticide use. NPJ Parkinsons Dis 10, (2024).
- Young, A. I., Wauthier, F. & Donnelly, P. Multiple novel gene-by-environment interactions modify the effect of FTO variants on body mass index. Nat Commun 7, (2016).
- Abdellaoui, A., Yengo, L., Verweij, K. J. H. & Visscher, P. M. 15 years of GWAS discovery: Realizing the promise. American Journal of Human Genetics vol. 110 179–194 Preprint at https://doi.org/10.1016/j.ajhg.2022.12.011 (2023).
- Sadee, W. et al. Missing heritability of common diseases and treatments outside the protein-coding exome. Human Genetics vol. 133 1199–1215 Preprint at https://doi.org/10.1007/s00439-014-1476-7 (2014).
- Qi, T., Song, L., Guo, Y., Chen, C. & Yang, J. From genetic associations to genes: methods, applications, and challenges. Trends in Genetics Preprint at https://doi.org/10.1016/j.tig.2024.04.008 (2024).
- Nam, Y. et al. Harnessing Artificial Intelligence in Multimodal Omics Data Integration: Paving the Path for the Next Frontier in Precision Medicine. Annual Review of Biomedical Data Science Downloaded from www.annualreviews.org. Guest (2024) doi:10.1146/annurev-biodatasci-102523.
- Shuni, W. X. Y. Q. Z. S. M. Gene–environment interaction analysis via deep learning. Genetic Epidemiology (2023).
- Mills, M. C. & Rahal, C. The GWAS Diversity Monitor tracks diversity by disease in real time. Nat Genetdoi:10.5281/zenodo.3600471.
- Martin, A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet51, 584–591 (2019).
- Westerman, K. E. & Sofer, T. Many roads to a gene-environment interaction. American Journal of Human Genetics vol. 111 626–635 Preprint at https://doi.org/10.1016/j.ajhg.2024.03.002 (2024).
- Genovese, G. et al. A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney Int 78, 698–704 (2010).
- Cohen, J. et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 37, 161–165 (2005).
- Krassowski, M., Das, V., Sahu, S. K. & Misra, B. B. State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Frontiers in Genetics vol. 11 Preprint at https://doi.org/10.3389/fgene.2020.610798 (2020).
- Wekesa, J. S. & Kimwele, M. A review of multi-omics data integration through deep learning approaches for disease diagnosis, prognosis, and treatment. Frontiers in Genetics vol. 14 Preprint at https://doi.org/10.3389/fgene.2023.1199087 (2023).
- Spanbauer, C. & Sparapani, R. Nonparametric machine learning for precision medicine with longitudinal clinical trials and Bayesian additive regression trees with mixed models. Stat Med 40, 2665–2691 (2021).
- The International Warfarin Pharmacogenetics Consortium. Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. New England Journal of Medicine 360, 753–764 (2009).
- Olopade, O. I., Grushko, T. A., Nanda, R. & Huo, D. Advances in breast cancer: Pathways to personalized medicine. Clinical Cancer Research vol. 14 7988–7999 Preprint at https://doi.org/10.1158/1078-0432.CCR-08-1211 (2008).
Follow the Topic
-
Human Genomics

Human Genomics is a peer-reviewed, open access journal that focuses on the application of genomic analysis in all aspects of human health and disease, as well as genomic analysis of drug efficacy and safety, and comparative genomics.
Related Collections
With Collections, you can get published faster and increase your visibility.
Artificial Intelligence in Omics and Translational Research
This special Collection in Human Genomics focuses on the integration of artificial intelligence (AI), machine learning (ML), and deep learning methodologies with omics and multi-omics data, with a particular emphasis on human health and disease modeling. As AI technologies rapidly evolve, their potential to revolutionize omics research and precision medicine is - especially in understanding genetic variation in human populations and advancing disease modeling - is becoming increasingly clear. However, bridging the gap between omics experts and AI specialists remains crucial for fully harnessing this potential.
We invite manuscripts featuring original studies that utilize state-of-the-art AI and deep learning algorithms applied to clinical, population-based, or other human-focused omics and multi-omics datasets. Contributions should demonstrate clearly how these methodologies can identify biomarkers, elucidate complex disease mechanisms, and inform precision medicine strategies, presented in a way accessible to researchers without extensive computational backgrounds. Additionally, this Collection aims to inform AI and computer science communities about the types and availability of omics datasets, with a focus on human-centric applications, thereby promoting interdisciplinary collaborations. Submissions that provide practical guidance, review emerging AI methodologies, offer clear tutorials, or highlight successful interdisciplinary case studies are also highly encouraged.
Together, we seek to enhance AI literacy within the omics community and foster collaborative innovation, accelerating translational discoveries and advancing human genomic applications with tangible impact on public health. Topics of interest include but are not limited to:
• Original research applying deep learning algorithms to human-focused omics and multi-omics datasets
• Reviews highlighting the current state-of-the-art AI methodologies, including generative and foundational models, and their applications in human disease modeling
• Tutorials introducing deep and graph-based learning concepts tailored for omics researchers
• Descriptions and characterizations of publicly available omics datasets suitable for AI applications, with emphasis on clinical or population-based datasets
• Perspectives and case studies on interdisciplinary collaboration between omics scientists and AI specialists
We encourage clear, concise, and accessible writing for interdisciplinary readers to support greater integration across these rapidly advancing fields.
This Collection supports and amplifies research related to SDG 3, Good Health and Well-Being .
Publishing Model: Open Access
Deadline: Jul 15, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in