Unveiling the Secrets of Soil Viromes: Biogeographic Patterns and Their Drivers Explored
Published in Earth & Environment and Microbiology

Viruses play a pivotal role in soil ecosystem functions by influencing host community composition and function through mechanisms like cell lysis, horizontal gene transfer, and host metabolism reprogramming. Despite their significance, the biogeography of soil viruses has not been as extensively studied as that of soil eukaryotic and prokaryotic life. This gap in knowledge hinders the enhancement of Earth systems and climate models and the development of effective natural resource management strategies.
In contrast to marine systems, where viral communities show relatively consistent patterns due to uniform environmental conditions, soil viruses are subjected to more extreme environmental variations and limited dispersal opportunities. This results in unique and complex interactions among soil viruses and a greater genetic divergence compared to marine systems. Previous studies have identified correlations between viral abundance and environmental factors such as soil water availability, temperature, and pH, but these have often focused on small, localized areas.
In the pioneering realms of soil virology, our research has embarked on an unprecedented journey to unveil the vast and uncharted diversity of soil viruses globally. The inception of the Global Soil Virome (GSV) dataset stands as a cornerstone of our endeavors, meticulously compiled from a comprehensive analysis of 1,824 soil metagenomes. This monumental dataset, aggregating approximately 30 terabytes of sequencing data across six continents, provides a panoramic view of the soil viral universe, spanning an eclectic mix of biomes and environmental conditions. The GSV illuminates the existence of 80,750 viral operational taxonomic units (vOTUs), a testament to the vast, yet predominantly uncharacterized, viral diversity beneath our feet. Despite the vastness of our dataset, a mere 3.3% of these vOTUs could be assigned to known viral families, underscoring the enigmatic nature of soil viral communities and the immense scope for discovery.

Metagenomics sheds light on uncovering the hidden diversity of soil viruses.
Our exploration extends beyond cataloging, delving into the intricate interactions between viruses and their hosts, with a special focus on predicting virus-host associations. The geographical and biome-specific patterns of viral diversity, illuminated by our research, underscore the profound influence of environmental factors on the assembly and distribution of viral communities. Our findings not only chart new territories in the understanding of soil viral biogeography but also underscore the intricate interplay between viruses, their hosts, and the environment, paving the way for a deeper understanding of soil ecosystem dynamics and the role of viruses therein. And our research is enriched by the creation of an interactive digital map and predictive biogeographic patterns (Figure 2), providing a multifaceted view of soil viral diversity. The interactive map allows users to visually explore the global distribution of viruses, enhancing understanding through direct engagement with the data. Simultaneously, predictive models, utilizing environmental datasets, reveal complex biogeographic patterns, offering insights into the factors driving soil viral diversity across different regions and biomes. These digital tools not only make our findings accessible but also enable a deeper comprehension of the underlying dynamics shaping soil viral communities globally.
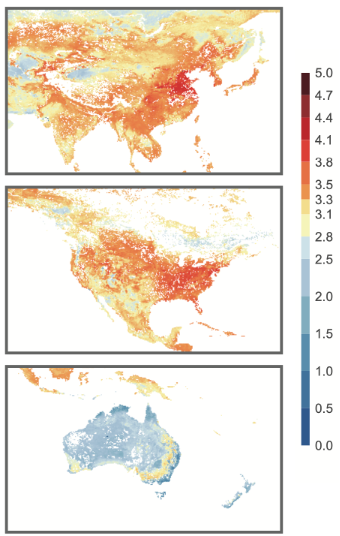
In this research, we reveal for the first time the ecological distribution of soil viral diversity across the globe, offering fresh insights into the intricacies of microbial existence within soil ecosystems. The creation of a comprehensive global soil virus database not only broadens our grasp of the microbial diversity of our planet but also underscores the critical role viruses may play in shaping the functions of soil microbial communities and their contribution to biogeochemical cycles. Despite the strides made, a vast expanse of soil viral "dark matter" remains uncharted, with countless virus species and their ecological roles still cloaked in mystery. Moving forward, it will be imperative to employ more harmonized and standardized approaches, coupled with in-depth functional analyses, to delve into these unexplored domains. These endeavors strive to deepen our comprehension of how soil viruses influence the vitality of ecosystems and agricultural yield, aiming to furnish scientific underpinnings for the stewardship and sustainable progression of global ecosystems.
Detailed information can be acquired through https://microbma.github.io/project/gsv.html and https://bmalab.shinyapps.io/global_soil_viromes.
Acknowledgements
We thank C. Kelly, C. Averill, D. Buckley, D. Goodheart, D. Duncan, D. Myrold, E. Eloe-Fadrosh, E. Brodie, E. Högfors-Rönnholm, H. Cadillo-Quiroz, J. Tiedje, J. Jansson, J. Norton, J. Blanchard, J. Schweitzer, J. Banfield, J. Gladden, J. Raff, K. Peay, K. Gravuer, K. M. DeAngelis, L. Meredith, M. Kalyuzhnaya, M. Waldrop, N. Fierer, P. Dijkstra, P. Baldrian, S. Theroux, S. Tringe, T. Woyke, T. Whitman, W. Mohn & San Diego State University for their permission to use their metagenome data.
Follow the Topic
-
Nature Ecology & Evolution

This journal is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biodiversity and ecosystem functioning of global peatlands
Publishing Model: Hybrid
Deadline: Jul 27, 2026
Understanding species redistributions under global climate change
Publishing Model: Hybrid
Deadline: Jun 30, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in