Using CRFasRNNs in Brain Extraction
Published in Computational Sciences and General & Internal Medicine
Introduction
Non-brain tissues can often share a similar MR signal intensity as brain tissues making statistical comparisons of brain data a hard process. Thus, brain segmentation is an inevitable step in almost every processing pipeline of brain MRI. Like every other image processing method the emergence of Deep Learning (DL) has rapidly advanced the brain extraction field. However, a lot of the innovations were based on using an abundant training dataset[1-3], whether multimodal or synthetic, and not the architecture itself.
Conditional Random Fields (CRF) [4] has been used as a post processing step of image segmentation methods widely before DL was introduced [5]. Then, CRFasRNN [6] was developed. The original CRF used in segmentation converges to an optimal segmentation output through iteration. CRFasRNN proposes to increase the flexibility and decrease time complexity of this by integrating the iteration to a Recurrent Neural Network.
There were attempts to use CRFs in volumetric medical imaging as well [7]. However, most of the methods used naive CRFs after the DL model architecture as a post processing step. A few researchers have attempted to use CRFasRNN in 3D medical images [8], but without any meaningful changes.
Our approach
We suspected the ineffectiveness of CRF to be from an imbalance in the features. Let us look at the fundamental equation from the CRF kernels used for 2D RGB images [5].

In the equation, p is positional information, I is the intensity information and w and 𝞡 control the weights of each features. Notice that there are two kernels, where each uses either the positional and intensity information (appearance) and only positional information (smoothness).
In 2D RGB images, we have 2 dimensions of positional features and 3 dimensions of intensity features. Thus, the equation can be overall balanced with the two kernels in the equation. However, in volumetric medical imaging, it is often the case that we have a grayscale image. Instead, we would have 3 positional features with 1 intensity feature.
Therefore, to effectively use the CRF layer with volumetric gray scale images, we investigate increasing the range of non-positional features.

Notice that we replace the image intensity I by c, which are features selected from the model architecture itself. More specifically, we select the features from the last layer of the model’s architecture before reducing the scale. This can tremendously increase the feature space and also pass more critical information than the image intensity itself, since it would have extracted specific features important for segmentation.
Additionally, we include a negative Dice Loss to further strengthen the CRF layer's effect on the prediction of the model. This negative Dice Loss is calculated between the prediction of the base model and the CRF layer's output. Thus, it serves as a regularizer enforcing the CRF layer to play a role in creating the final prediction. The figure below provides a summary of the proposed architecture.
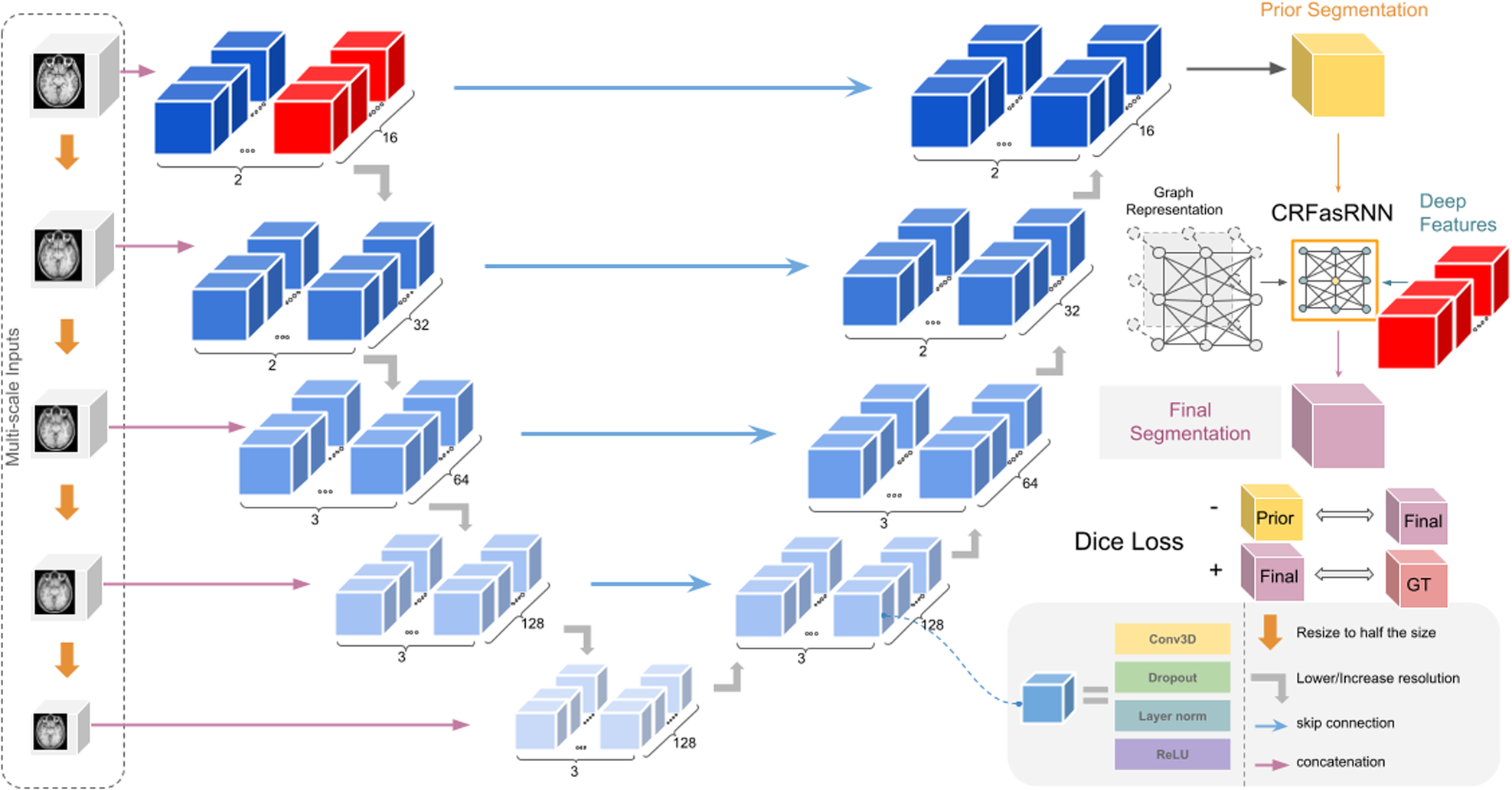
Results
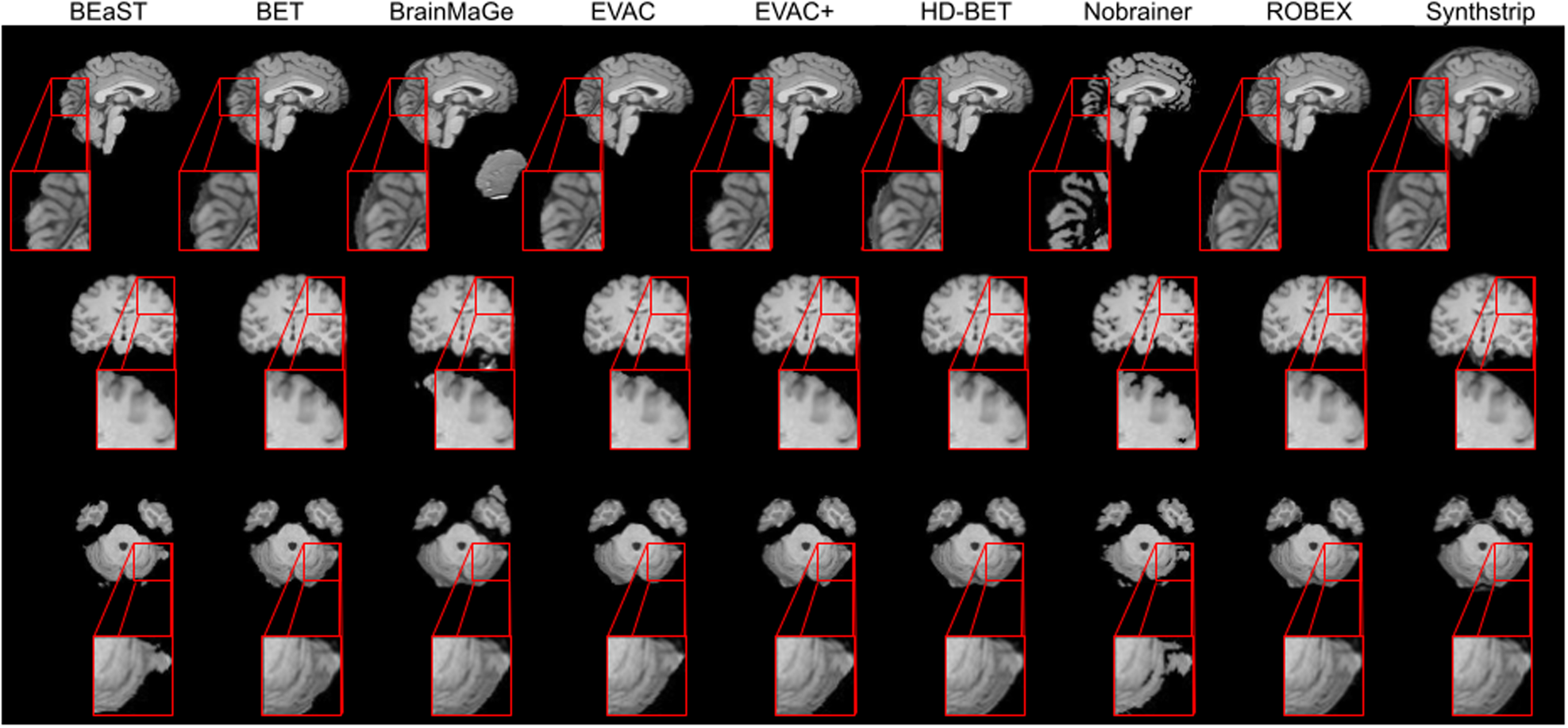
Results show that our approach can achieve competitive segmentation near the boundaries. It is worth emphasizing that this accuracy is from training only on public datasets where a large portion did not have manual annotations.
Conclusion
Our approach revisits CRFs as a tool in volumetric medical image segmentation, and we find that contrary to prior beliefs, CRFasRNN can be useful in such tasks if calibrated. Currently a student model of the method is available in DIPY [9] for easier use. The full method is publicly available as well and the code can be found in the link below.
https://github.com/pjsjongsung/EVAC
1. Thakur, Siddhesh, et al. "Brain extraction on MRI scans in presence of diffuse glioma: Multi-institutional performance evaluation of deep learning methods and robust modality-agnostic training." Neuroimage 220 (2020): 117081.
2. Schell, M., et al. "Automated brain extraction of multi-sequence MRI using artificial neural networks." European Congress of Radiology-ECR 2019, 2019.
3. Billot, Benjamin, et al. "SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining." Medical image analysis 86 (2023): 102789.
4. Lafferty, John, Andrew McCallum, and Fernando CN Pereira. "Conditional random fields: Probabilistic models for segmenting and labeling sequence data." (2001).
5. Krähenbühl, Philipp, and Vladlen Koltun. "Efficient inference in fully connected crfs with gaussian edge potentials." Advances in neural information processing systems 24 (2011).
6. Zheng, Shuai, et al. "Conditional random fields as recurrent neural networks." Proceedings of the IEEE international conference on computer vision. 2015.
7. Kamnitsas, Konstantinos, et al. "Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation." Medical image analysis 36 (2017): 61-78.
8. Monteiro, Miguel, Mário AT Figueiredo, and Arlindo L. Oliveira. "Conditional random fields as recurrent neural networks for 3d medical imaging segmentation." arXiv preprint arXiv:1807.07464 (2018).
Follow the Topic
-
Communications Medicine

A selective open access journal from Nature Portfolio publishing high-quality research, reviews and commentary across all clinical, translational, and public health research fields.
Related Collections
With Collections, you can get published faster and increase your visibility.
Reproductive Health
Publishing Model: Hybrid
Deadline: Mar 30, 2026
Healthy Aging
Publishing Model: Open Access
Deadline: Jun 01, 2026
Latest Content
Why is Singapore Identified in Global Research as Number One? How Physical Activity and Education Excellence Created a Global Leader




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in