Viral channelrhodopsins
Published in Microbiology

Channelrhodopsins are microbial rhodopsins that directly convert absorbed light into ion fluxes across cellular membranes by opening conductive pores, and are widely used as optogenetic tools for manipulating neuronal activity. Although known to science for almost 20 years already, until recently these cation- or anion-conducting ion channels have been characterized exclusively from cultured unicellular algae. The last year has seen the discovery of several new families of these rhodopsins - in heterotrophic and photosynthetic eukaryotic microbes (e.g. the MerMAIDs [Oppermann et al. 2019] and channelrhodopsins from the marine stramenopiles Labyrinthulea and haptophyte algae [Govorunova et al. 2020]).
To search for channelrhodopsins with interesting activities and in general to better understand their diversity, we look for channelrhodopsin sequences from different metagenomic datasets with special emphasis on the data generated as part of the Tara Oceans project. Using this approach we previously discovered and characterized the MerMAID rhodopsins: anion-conducting channelrhodopsins with unique photocurrent properties. Along with the MerMAIDs, we unearthed (unoceaned) several rhodopsin sequences, suspected to be channelrhodopsins, unusual in a different way. These came from metagenomic contigs of suspected viral origin. This observation was of special interest as no one ever observed these genes in viruses before. While this is nice, in order to tell a story, a confirmed channeling activity and verification on the viral origin are needed.
Initially these channels gave only very tiny photocurrents, just above noise, which prevented us to conclude about any activity. It needed manipulation in the N-terminus part to improve expression in order to get a signal one can work with (see screen photo, top figure), and then it needed the truncation of the long non-membranal C-terminus part to get a real signal (see screen photo, bottom figure), increasing the photocurrent amplitude to about 800pA (-80mV)! This was performed exactly 2 years ago. But still most of the channels did not reach the cytoplasmic membranes and therefore there were inconsistencies with current measurements. It then took the long and painstaking path of rebuilding the constructs in order to confirm these initial observations to determine that these channels were indeed anion-conducting channelrhodopsins.
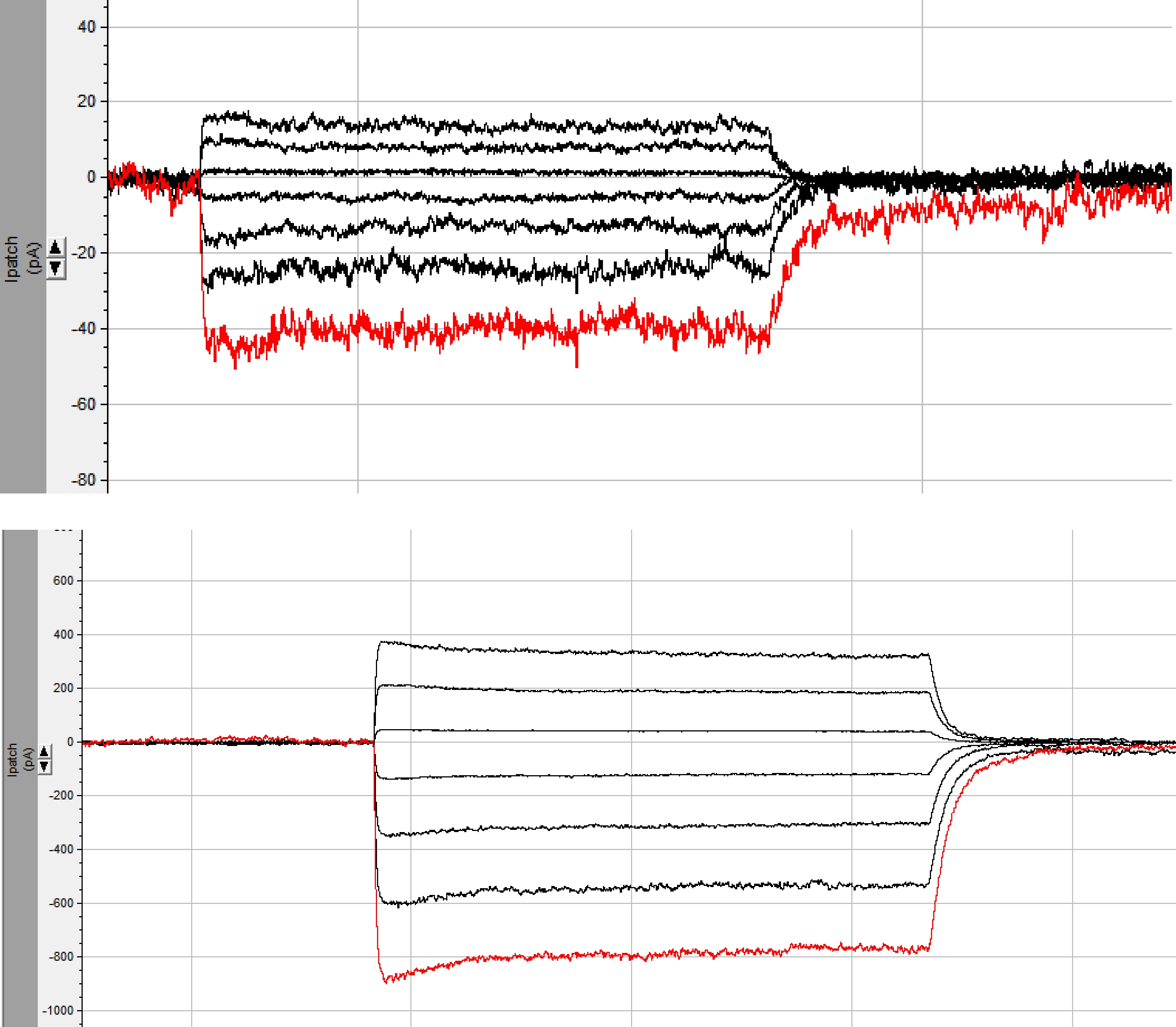
Screenshots of the initial raw photocurrents measured for one of the viral channelrhodopsins (pictured exactly two years ago). Top: measurement after addition of a chimeric N-terminus from ChR2/ChR1; Bottom: measurement after additional truncation of the C-terminus, leading to 20-fold increase of photocurrent amplitudes (compare different scales; photo credit: Jonas Wietek)
What was strange from the onset was the fact these channelrhodopsins were apparently present only in viruses. So, we decided to try harder and search for possible donors of the viral channelrhodopsins among cellular life. Thanks to two big repositories of transcriptomic data on various algae, we discovered, to our big surprise, that related proteins were present in some green algae. Surprising was the fact that green algae were already known to harbor channelrhodopsins of a different kind - the best characterized family of the green algal cation-conducting channelrhodopsins. Nevertheless, a peculiar picture started to emerge in which the “primitive” unicellular green algae (prasinophytes) appeared to possess multiple families of channelrhodopsins, one of which was the anion-conducting channelrhodopsins, and another one - putative cation-conducting channelrhodopsins. We noticed that the channelrhodopsins were present only in those prasinophytes that have eyespots, the light-sensitive organelles, indicating that they likely participate in vision. How the two families might cooperate remains a mystery. How can opening channels of apparently opposite action in response to the same light signal transform into a consistent cellular response? Perhaps the anion-conducting channelrhodopsins and the cation-conducting channelrhodopsins in prasinophytes do not open simultaneously, are expressed at different times, or have a different subcellular localization? This riddle will be solved in future experiments.
Now, what about those viruses with channelrhodopsin genes? It is not always easy to define the origin of the relatively short metagenomic fragments. To solve this problem we went out on a quest to find viruses in culture that might be related to our viral scaffold. It was clear that none of the viruses with complete genomes possessed channelrhodopsin genes and didn’t show close similarity to the viral fragments we collected. Phylogeny suggested that the channelrhodopsins originated from a particular group of green algae - those of Pyramimonas, peculiar scaly algae with many flagella. Analogously, there appeared to be a couple of other genes on the viral contigs that could be traced back to Pyramimonas as well. While many viruses are known to infect green algae, only one isolate has been described thus far from Pyramimonas - Pyramimonas orientalis virus 01B (PoV-01B, Mimiviridae). It was isolated in Norway some 20 years ago, but unfortunately was lost in culture (as often happens with marine viruses). Luckily not before some initial genetic data could be collected and some samples frozen for better times. Hoping to find the channelrhodopsin gene, we could not miss the opportunity and obtained a near-complete genome assembly for this virus. While to our amazement the PoV-01B genome indeed had a region of high similarity to one of the channelrhodopsin-possessing contigs, it didn’t contain the channelrhodopsin gene itself. Further analyses reinforced the idea of a common origin of some of the channelrhodopsin-possessing viruses with PoV-01B and other members of the mesomimiviruses, a group of giant viruses that mostly infect various unicellular algae. Curiously, another metagenomic fragment with a channelrhodopsin gene didn’t show any affinity to mesomimiviruses and seemed to be part of a different, remotely related group of algae-infecting giant viruses. The lack of the channelrhodopsin gene in PoV-01B forced us to put its viral origin into question. Perhaps we were simply dealing with a fragment of viral DNA that got fossilized in the genome of a green alga (as it happens) and later gave refuge to an algal channelrhodopsin gene? Analysis convincingly rejected this hypothesis: the channelrhodopsin genes were “domesticated” by the viruses a sufficiently long time ago and no traces of their algal origin could be detected on the DNA level.
Thus, we could conclude that the channelrhodopsins were acquired by viruses from their photosynthetic algal hosts and got fixed and maintained in some viral lineages for a period of time. Moreover, after hijacking one such gene, unrelated viruses shared it among themselves. The fact that not all of the corresponding viruses had this gene, must indicate that it is not required for survival of the virus. What benefits it might provide to the virus remains an even bigger question than the function of the anion-conducting channelrhodopsins in their putative hosts. As the algal proteins are likely implicated in behavioral responses, we can speculate that their viral homologs have a similar physiological role. It is possible that the viral anion-conducting channelrhodopsins cause a significant behavioral change in the algal host, advantageous for the residing virus.
Our findings show the power and potential of combining metagenomics and electrophysiology for the discovery of new rhodopsins. We are now in the process of searching more environmental datasets, on the DNA and RNA levels, in order to find new channelrhodopsins and novel biophysical properties.
We thank Richard Pienaar and Stuart D. Sym for providing us with the micrograph of viral infection in Pyramimonas pseudoparkeae (used for the poster image).





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in