When a positive interaction is bad: a link between mosquito-specific viruses and transmission of dengue and Zika virus
Published in Microbiology

Whenever you hear about viruses and mosquitoes together, you will probably think about arboviruses that cause human diseases, such as dengue and Zika. This association is understandable since dengue virus alone is estimated to cause about 400 million infections every year, and nearly 4 billion people are at risk of being infected by this virus. Climate change, globalization, urbanization and deforestation are worsening this scenario, increasing the geographical distribution of the main mosquito vectors for human viruses, Aedes aegypti and Aedes albopictus. Currently, no efficient vaccines or antiviral drugs are available for most mosquito-borne viruses, placing most efforts in vector control to minimize disease transmission. We would like to present here a new perspective on mosquito-virus interactions from which you will discover that the collection of viruses (i.e., the “virome”) infecting mosquitoes goes beyond arboviruses, and how these recently explored biological agents can help us answer relevant questions about the mosquito permissiveness and their ability to transmit viruses.
Our work started with a large-scale virological surveillance of Ae. aegypti and Ae. albopictus collected in six countries on four continents with the help of a network of collaborators, the majority from the ZIKAlliance consortium. Over 800 mosquitoes were pooled into 91 samples from which we generated small RNA libraries that were subjected to high throughput sequencing. From this moment, we started a long bioinformatics journey into this work.
In 2015, our group developed a sequence-independent virus characterization approach based on the small RNAs (usually from 18 to 30 nucleotides in length) generated by host-virus interactions. In this work we expanded our efforts to characterize viruses in field mosquito samples. Our small RNA-based metagenomics approach allowed us to investigate special characteristics of small RNAs generated during virus infection, used here to overcome the challenge of differentiating exogenous from endogenous viral sequences from eukaryotic metatranscriptomes. These characteristics also allowed us to infer active virus replication based on the type of small RNA profile generated by each virus-derived sequence, be it endogenous or exogenous.
In this work, our purpose was to provide a reliable and curated list of viruses circulating in the two most widespread vector mosquito species. We were not only worried about generating a list of viruses but also trying to better understand the phenomenon of concomitant viral infections in Aedes mosquitoes. We obtained a list of 12 unique viruses from our 91 samples, five of which were potentially new viral species. All 12 viruses are presumed insect-specific viruses (ISVs), meaning that they cannot infect vertebrates and two of them, Phasi Charoen-like virus (PCLV) and Humaita Tubiacanga virus (HTV), caught our attention due to their high prevalence in Ae. aegypti worldwide. Using a spatiotemporal analysis of field samples, we observed that PCLV and HTV were positively associated with dengue virus infection in mosquitoes. We further investigated interaction and showed, in controlled laboratory conditions, that HTV and PCLV rendered Ae. aegypti more susceptible to dengue and Zika viruses. Moreover, we observed that coinfection between HTV and PCLV together with ZIKV shortened the time necessary for the mosquito to transmit the virus, also known as extrinsic incubation period (EIP). Mathematical modeling showed that shortening the EIP by one or few days could have a significant impact in the number of human cases in the population.
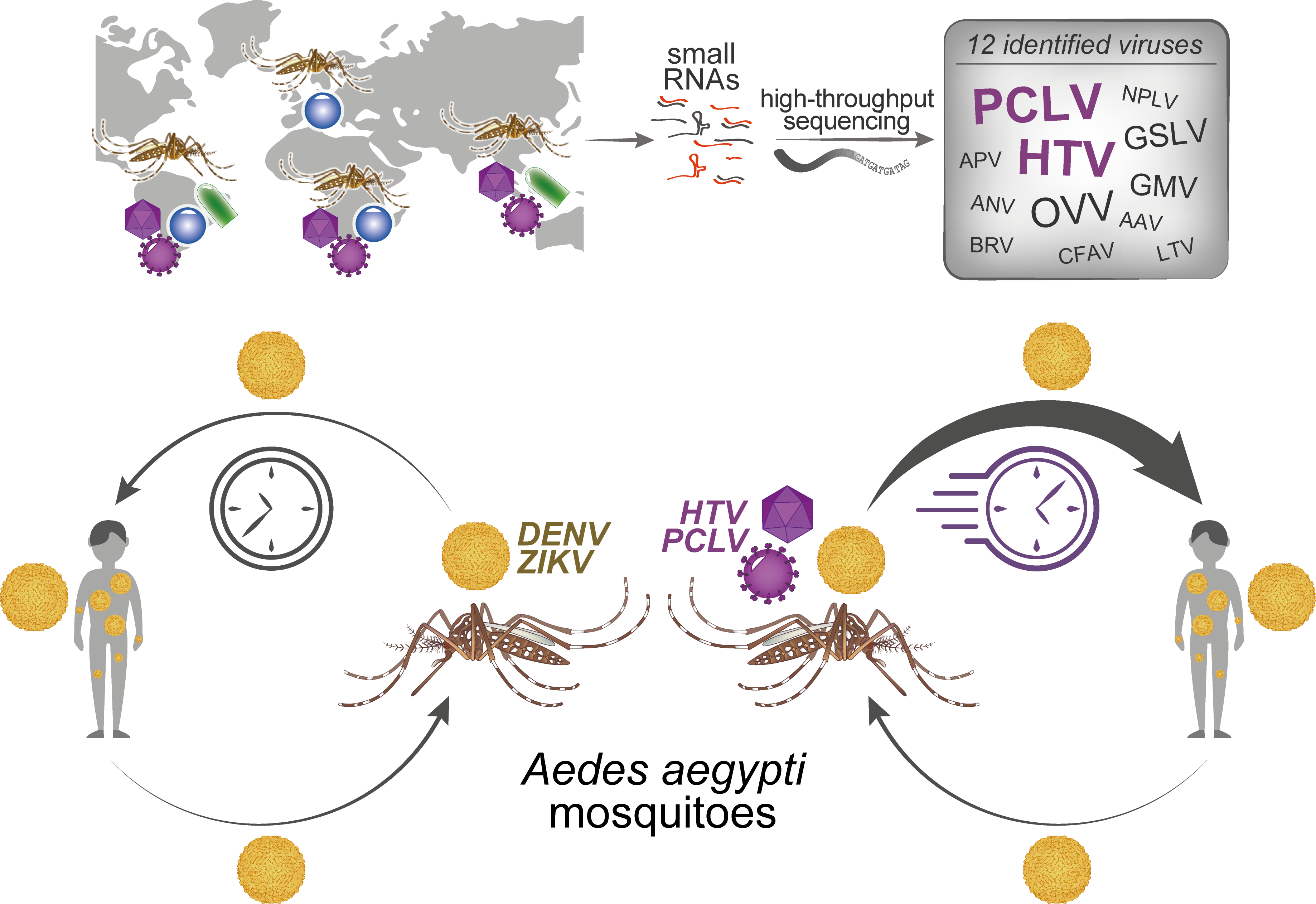
In this work we investigated the virome (i.e., virus repertoire) of Ae. aegypti and Ae. albopicus mosquitoes from 4 continents and identified 12 circulating viruses. Strikingly, two insect-specific viruses were present throughout the globe, Humaita-Tubiacanga virus (HTV) and Phasi Charoen-like virus (PCLV), and prompted the question if they could affect arbovirus transmission by the mosquito. We observed that, both in Ae. aegypti natural populations and artificially infected laboratory mosquitoes, HTV and PCLV interacted positively with dengue virus (DENV) and Zika virus (ZIKV). This interaction led to higher DENV and ZIKV loads, accelerating virus transmission from the mosquito to a mammalian host and potentially impacting the progression of Aedes-borne disease outbreaks.
At this point, we raised an important question: how do HTV and PCLV affect arbovirus infection in Ae. aegypti? Again, we resorted to a high-throughput approach and performed genome-wide transcriptomic analysis. Our results suggested a link between the expression of histone genes and the observed positive interaction between viruses. Here began new challenges for our bioinformatics team. We were going after genes usually organized in genomic clusters with multiple copies and complex regulation of translation. Through a series of experiments involving gene silencing and virus transmission, we demonstrated that HTV and PCLV prevented downregulation of histone H4, a previously unrecognized pro-viral host factor in mosquitoes, leading to increase DENV and ZIKV transmission to a vertebrate host.
Our work highlights how important it is to evaluate ISV circulation in vector insects as a parameter for risk analysis of outbreaks. Mosquitoes often can carry productive virus infections throughout their life with minor fitness costs, and we are at early stages of understanding the impact of multiple viral coinfections in arbovirus transmission. Moreover, these interactions could help understanding the differences in vector competence (i.e., capacity of transmitting a virus) between species such as Ae. aegypti and Ae. albopictus. There are many discussions about the fact that Ae. albopictus naturally harbors the bacteria endosymbiont Wolbachia and its influence on arbovirus by this species. Interestingly, among the 12 identified viruses in our study, 10 were found in A. aegypti and only 2 in A. albopictus, suggesting a less diverse virome in the latter. The diversity of ISVs offers another point of view for understanding the differences in the susceptibility to arboviruses among mosquito species.
Furthermore, many ISVs and arboviruses have close phylogenetic proximity, and it is likely that arboviruses have evolved from ancestral ISVs that acquired the ability to “jump” to the vertebrate hosts that their primary hematophagous vectors fed on. Further studies about the biology of ISVs are important to shed light on the origin and evolution of arboviruses, raising preparedness before the emergence of new threats to human and general animal health.
This is a brave new world of mosquito-virus interactions that will benefit from a systemic approach integrating the virome, transcriptome, genome, and environmental factors to understand arbovirus transmission by the insect vector. We are glad to be navigating this untamed field, combining classical bench-work with bioinformatics and system biology to discover the repertoire of viruses in insects and reveal their interactions with each host.
Access the full paper here.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in