When DNA words started to repeat and took over the world!
Published in Ecology & Evolution and Genetics & Genomics

The Beginning
Almost six years ago, our new evolutionary genomics lab at the Indian Institute of Science Educaton and Research, Bhopal began exploring the impact of gene loss on organisms' evolution and adaptation. In our initial study of the CYP8B1, we discovered that gene loss is primarily preceded by the relaxation of purifying selection [1]. This laid the foundation for our subsequent research. As we delved deeper into the realm of relaxed selection, gene loss, and evolution, we observed that the intricate interplay of these factors helps organisms adapt to specific environmental demands. We extensively studied this phenomenon in the PLGRKT [2].
Innovations
The exploration of gene loss using next-generation data required clever and definitive methods to identify gene loss across various vertebrate species. We developed a five-pass strategy to identify gene loss and its role in organisms' evolution (Figure 1).
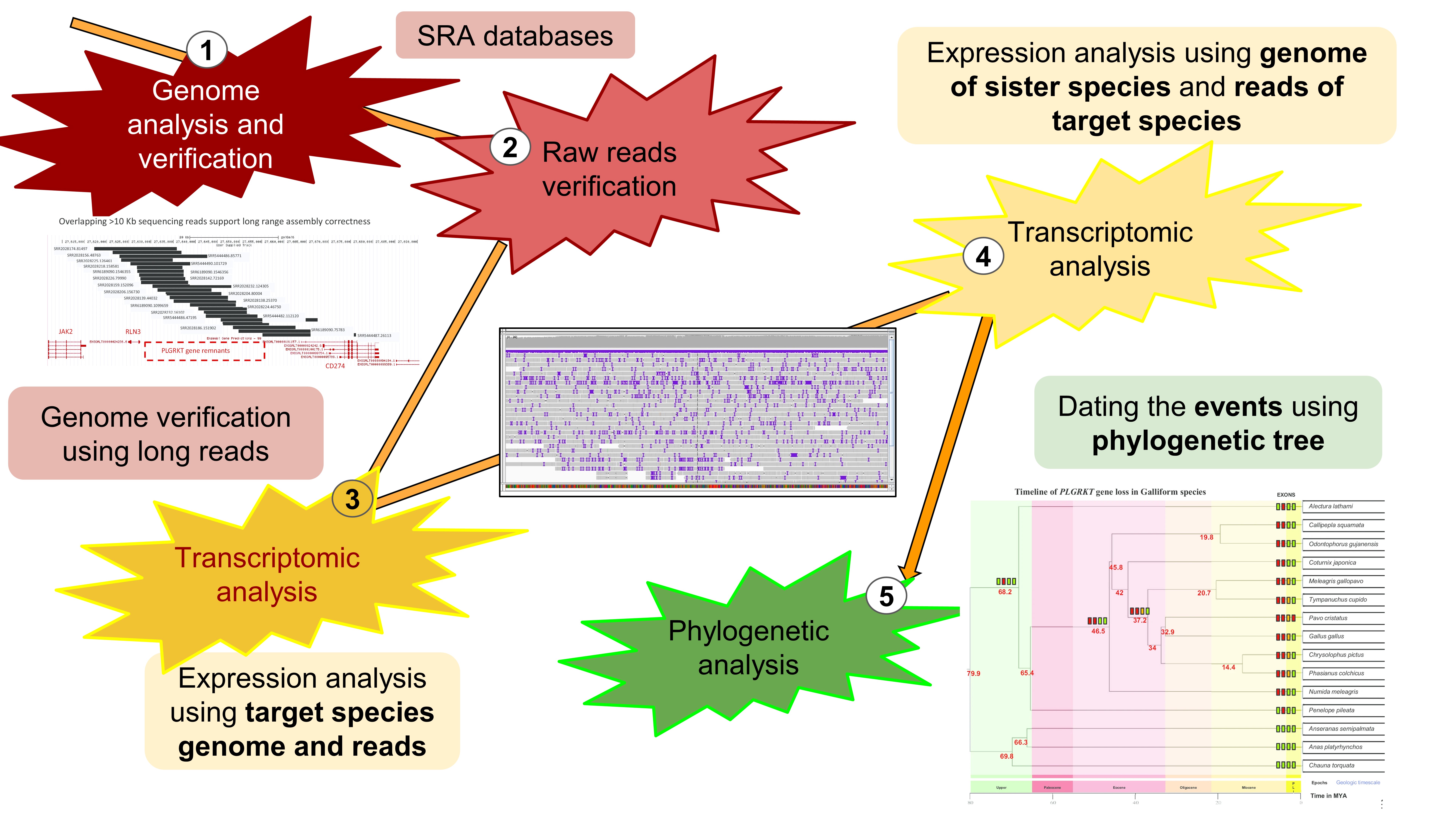
Linking Loss and Function
Building on our previous studies, we established the essential role of gene loss and relaxed selection in evolution. We aimed to extend and link this phenomenon with its consequences on morphology and function. This led us to investigate COA1 loss in vertebrates [3]. Here, we identified a significant link between the loss of flight in birds and the loss of the COA1 (Figure 2).
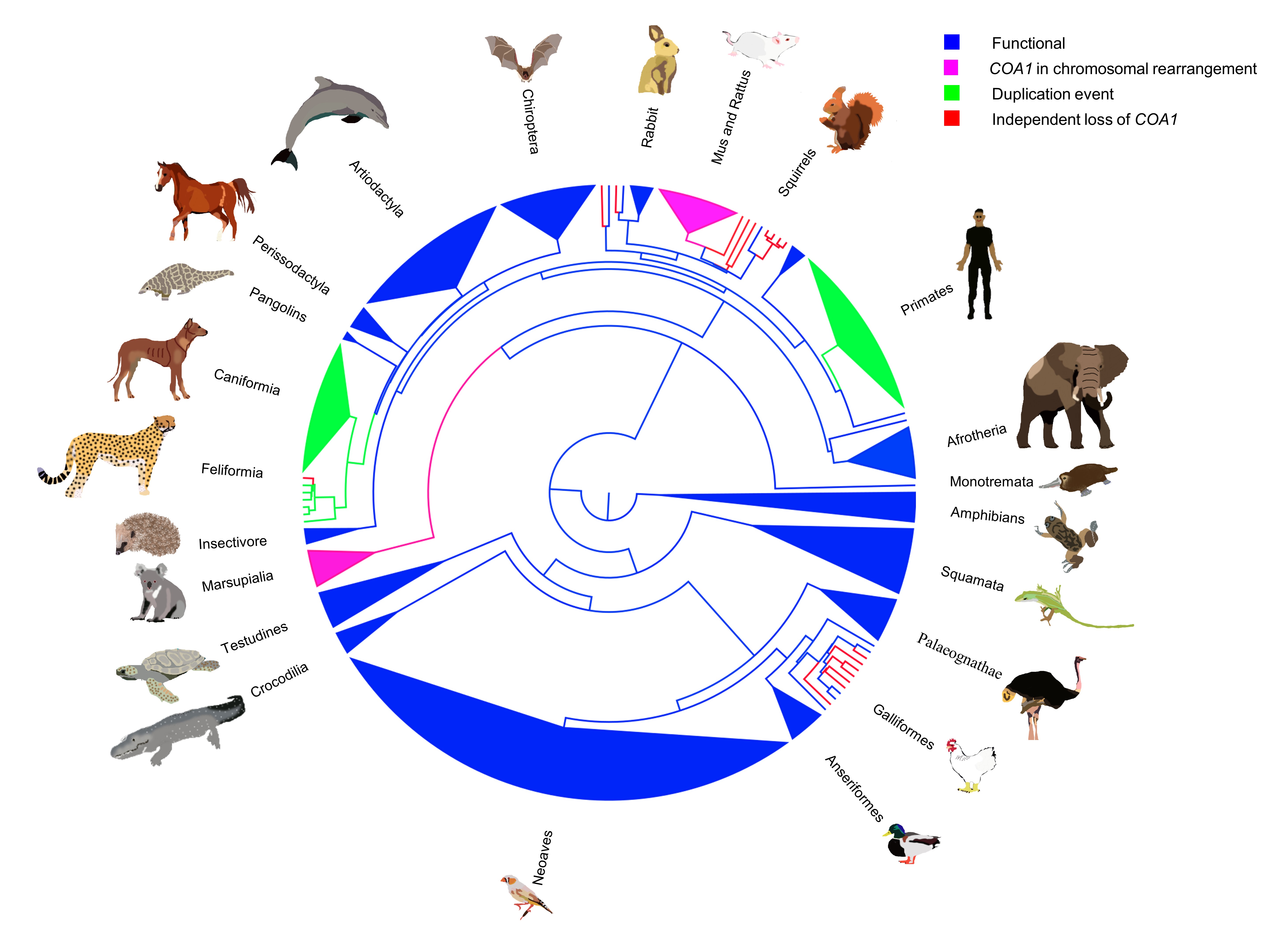
Exploring New Horizons
Having explored gene loss, relaxed selection, and their effects on function, we turned our attention to another phenomenon in genome evolution—amino acid repeat variability. This phenomenon, around 100,000 times more prominent than point mutations, became the focus of our study on immune genes. We examined the effects of amino acid repeats on morphological variations and evolutionary novelties [4].
New Challenges: New Innovations
While detecting general patterns of amino acid repeat distribution and types was straightforward, exploring variability required innovative strategies to empirically quantify length contrasts between species. We employed phylogenetically independent contrasts (PIC) and statistical tests for this purpose. This approach helped us identify candidate genes for future exploration (Figure 3).
Taking Flight with Birds
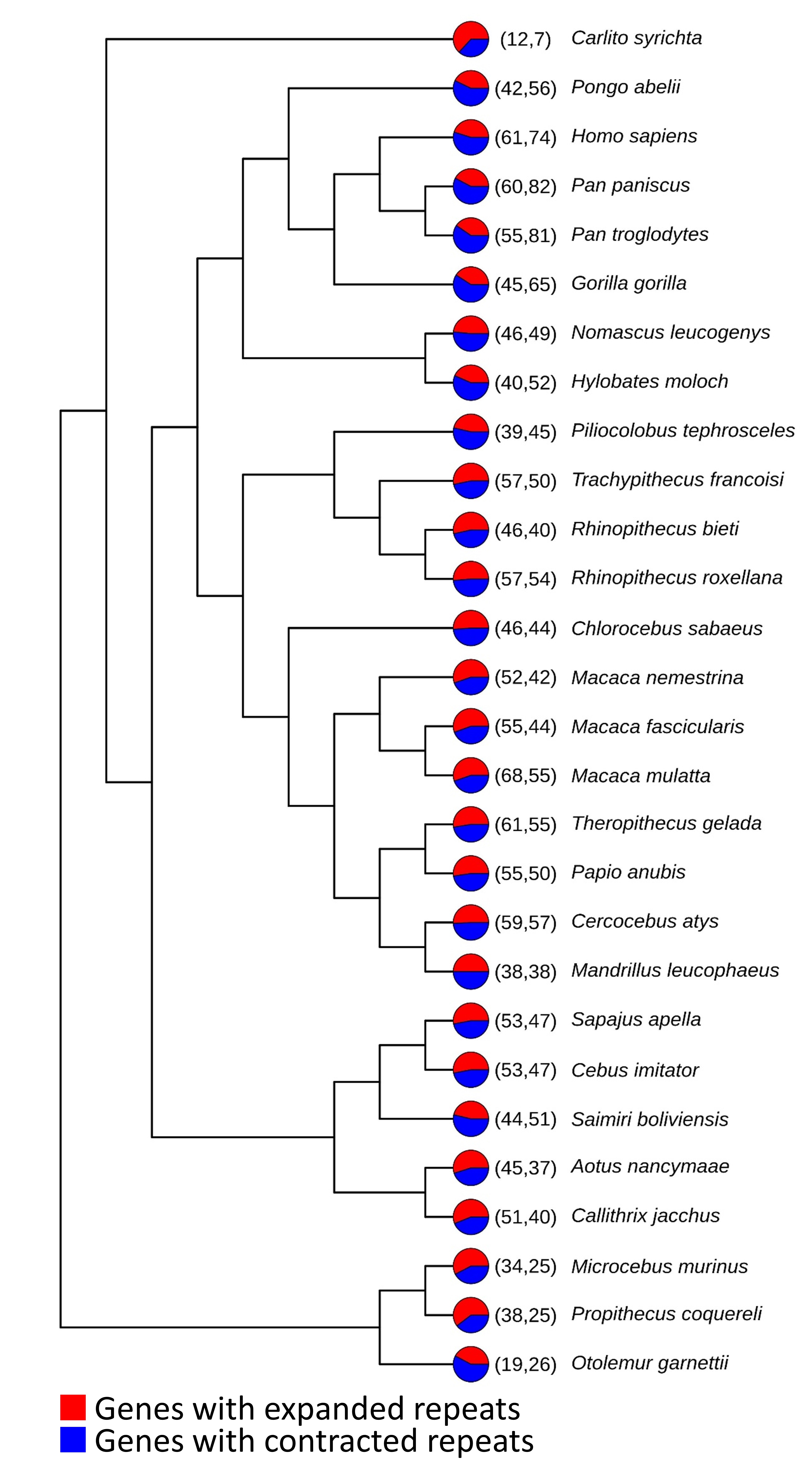
In our research journey, we utilized our accumulated knowledge to explore the fascinating group of species, Aves (Figure 4).
Having gained insights into the role of amino acid repeats in providing evolutionary innovations, we focused on birds as the most diverse and species-rich clade (Figure 5).

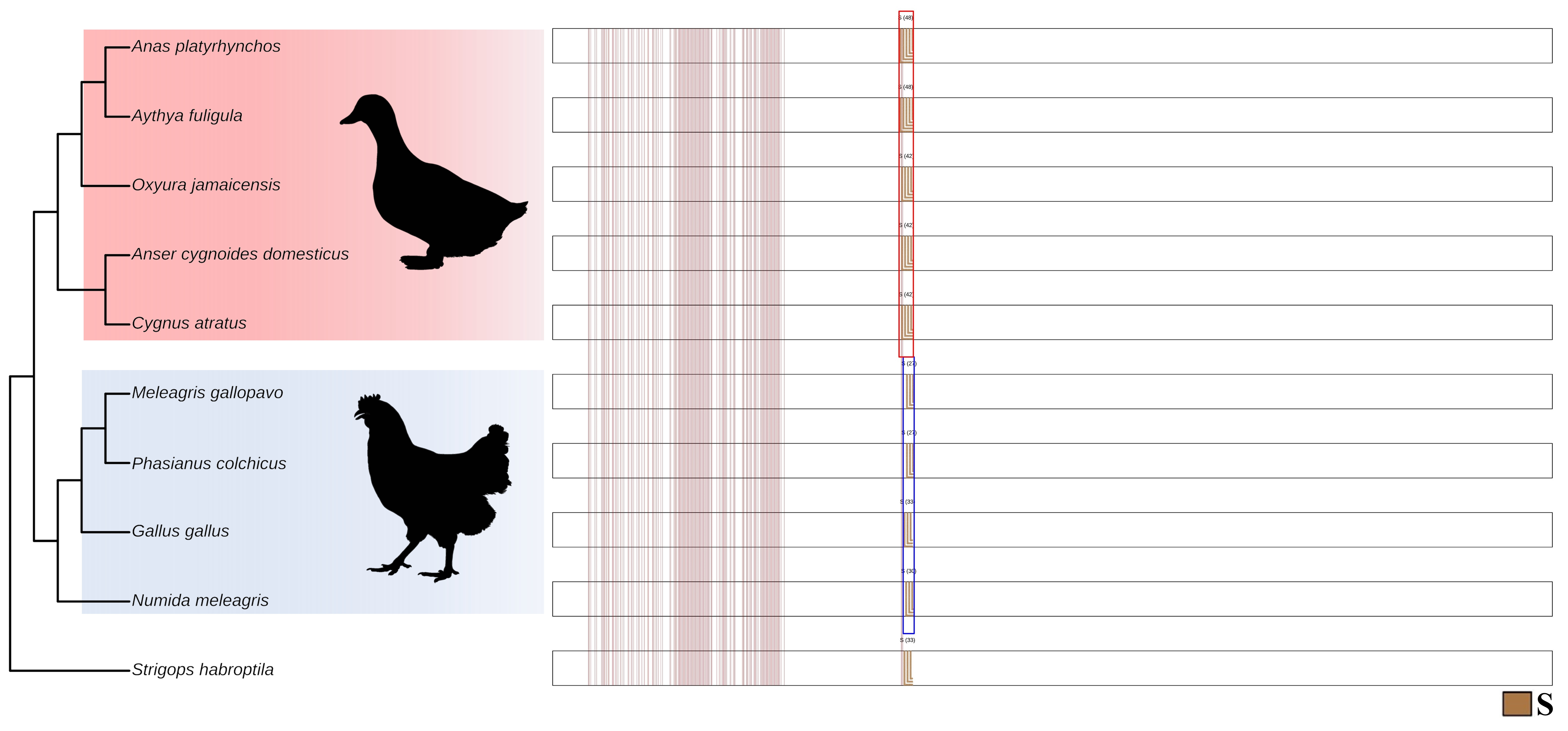
This project was particularly exciting for us, and our findings motivated us to continue. We identified overall patterns of length expansion and contraction of amino acid repeats in each species/clade along the bird phylogeny [5]. Additionally, we identified the PCDH20 with polyS variability between waterfowls and landfowls, potentially connecting differences in hearing abilities to their environment (Figure 6).
What’s Next?
Our journey continues on this exciting path as we explore the role of amino acid repeats in all protein-coding genes across the available high-quality genomes of Tetrapoda species [6].
After graduating, I (Sandhya Sharma) am planning to apply for a post-doc position internationally so I can continue to carry out research.
References
- Shinde S.S., Teekas L., Sharma S., Vijay N. 2019. Signatures of Relaxed Selection in the CYP8B1 Gene of Birds and Mammals. J. Mol. Evol. 87, 209–20.
- Sharma S., Shinde S.S., Teekas L., Vijay N. 2020. Evidence for the loss of plasminogen receptor KT gene in chicken. Immunogenetics. 72, 507–15.
- Shinde S.S., Sharma S., Teekas L., Sharma A., Vijay N. 2021. Recurrent erosion of COA1/MITRAC15 exemplifies conditional gene dispensability in oxidative phosphorylation. Sci. Rep. 11, 24437.
- Teekas L., Sharma S., Vijay N. 2022. Lineage-specific protein repeat expansions and contractions reveal malleable regions of immune genes. Genes Immun. 1–17.
- Sharma S., Teekas L., Vijay N. 2023. Protein Repeats Show Clade-Specific Volatility in Aves. Mol. Biol. 57, 1199–211.
- Teekas L., Sharma S., Vijay N. 2023. Terminal regions of a protein are a hotspot for low complexity regions (LCRs) and selection. bioRxiv. 2023.07.05.547895.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in