
Background
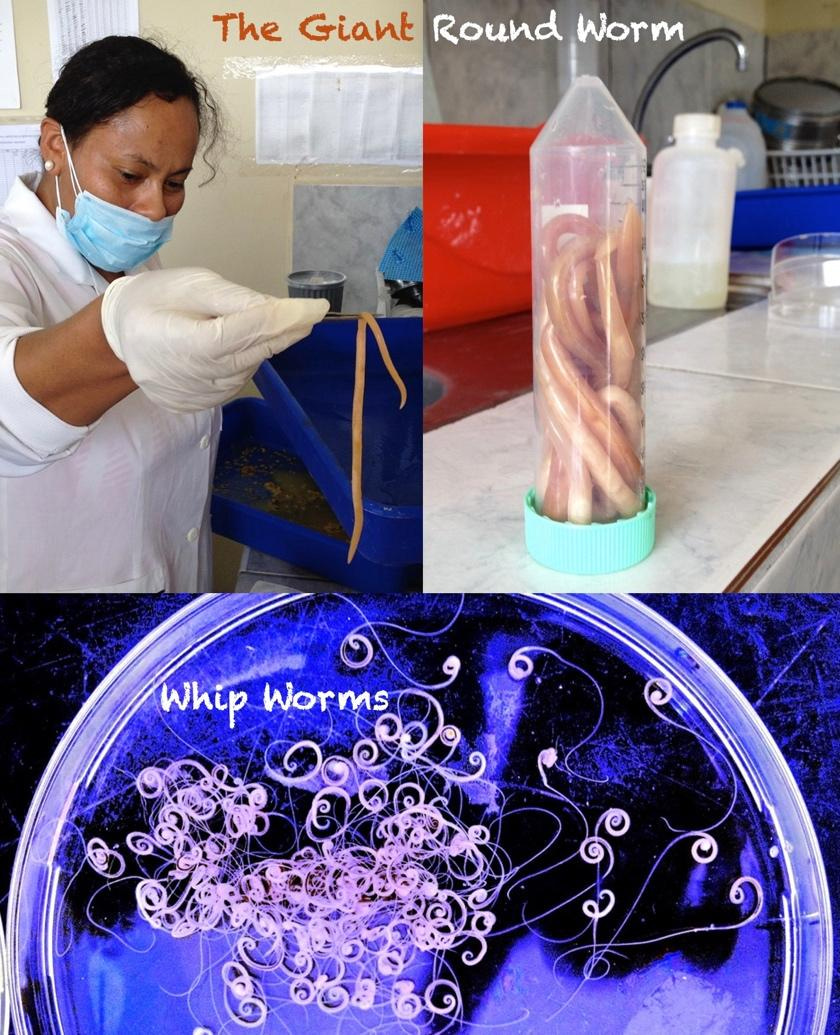
Nematode worms are one of the most successful and abundant organisms on Earth, being found on every continent and ecological niche. Several of these worms have evolved a parasitic lifestyle where their abundance persists afllicting a quarter of humanity, as well as billions of animals infected in the agriculture industry and our pets. Although widespread their impact is often relatively benign, chronic and persistent, yet a few species cause serious diseases. Notable examples include the filarial nematodes (causative agents of elephantiasis, river blindness and heartworm), the hookworms, whip worms and giant round worms (see Figure 1).
Parasitologists have studied these nematodes for hundreds of years trying to unravel the complex biology concealed within a simple tube-like structure. For the past 25 years, the Filariasis Lab at The Liverpool School of Tropical Medicine has been dissecting the role of a bacterial endosymbiont of filarial nematodes, called Wolbachia. Dr Shannon Quek, lead author of the Nature Microbiology paper, carried out his PhD on this bacterial endosymbiont’s impact on parasite host transcriptomes and discovered their essential role in the transmission of the parasite through its blood feeding mosquito vector (DOI: 10.1073/pnas.2120003119). Following his PhD, he expanded his work on Wolbachia into mosquitoes in Grant Hughes’ lab, where he also started to look how to discover another important microbiome of mosquitoes – viruses.
Serendipity Part One
My (Shannon’s) ‘light-bulb’ event that triggered this new discovery was to apply the mining tools used to probe mosquito viromes and apply them to parasitic nematodes. I started my screening looking at Brugia malayi, the subject of my PhD. Much to my surprise the first trawl of the worm’s transcriptome revealed a hidden viral genome – BMRV1. Worried that it might just be a contaminant from the parasite’s host or mosquito vector, I checked other transcriptomes, as well as stocks of nucleic acids from different sources of the same lab strain and found the virus in all different isolates. So, on top of its bacterial endosymbiont, the Brugia parasite appeared to have a viral symbiont in its microbiome.
Now confident in my discovery, I broke the news to the head of the lab, Prof Mark Taylor, who was just as surprised as I was and both excited and impressed by my discovery. Next in the queue was Onchocerca volvulus, another filarial nematode, which causes skin disease and blindness primarily in adults in Africa. Recently a new and serious disease presentation has been linked to onchocerciasis – Onchocerciasis-Associate-Epilepsy (OAE) - affecting children and adolescents in areas of high parasite transmission, known locally as ‘Nodding syndrome’ and ‘Nakalanga syndrome’. We were aware that an OAE expert in Belgium, Prof Bob Colebunders, had been looking into what might be causing this serious paediatric epilepsy, and was currently focusing on possible viral pathogens in the worm or its blackfly vector. I started to look using my virome discovery algorithm and, to both of our surprise, I found another virus. This time, it was a Rhabdovirus, related to lyssaviruses, such as rabies, which cause fatal neurological disease - “I think we’d better have a chat with Bob!”
Serendipity Part Two
During our Teams call Bob called over his PhD student Amber Hadermann, to have a look at the new virus genome – “Yep, that’s the same one!” was Amber’s excited response. It turns out that their team started looking for viruses in O. volvulus after finding a multitude of them in its vector, the blackfly (DOI: 10.1101/2024.04.05.588247). Purely out of curiosity, they started doing metagenomics on the worm together with Prof. Jelle Matthijnssens at the Rega institute. During this technique, all potentially present viruses are sequenced after they are filtered from liquified worms. Just like Shannon, after analysing the data with PhD student Lander De Coninck, she immediately was interested in that one rhabdovirus and already started hypothesing its possible involvement in OAE. Though most of their team remained sceptical of their discovery, up until that one Teams call. From that point on, they were finally certain that they indeed discovered a rhabdovirus and started silently hoping that this might be the breakthrough for OAE they had all been working towards.
So, two labs looking with different data mining tools and using parasites and data sets from different endemic countries both converged on the same virus – OVRV1. Now started a journey to see how many other parasitic nematodes hosted a hidden virome. After churning through all publicly available transcriptomes from 41 species of parasitic nematodes, we discovered a grand total of 91 new viruses, infecting ~70% of the parasitic nematodes of humans and economically important animals, which was published in the October issue of Nature Microbiology (DOI: 10.1038/s41564-024-01796-6).
What these viruses are doing within these parasites and whether they contribute to parasite diseases are important research questions we are now starting to address. RNA viruses are also found in protozoan parasites and those in Leishmania parasites are known to drive the immune response away from the parasite leading to worse disease and parasite persistence (DOI: 10.1128/mbio.02642-22). We tested a large serum bank from onchocerciasis endemic areas and found nearly everyone made antibodies to the surface glycoprotein of OVRV1 – showing that the virus is released into the blood and tissues of the human host and is recognised by the immune system. So, more work needs to be done, but we now have a candidate virus, which might trigger the pathogenesis of Onchocerciasis-Associated Epilepsy. As for the 90 other viruses? – we are now linking up with and reaching out to parasitologists across the globe working on these nematode parasites to see what impact they might have on nematode biology, whether they are pathogenic to their nematode host and contribute to nematode diseases of people and animals.
If your parasite is on our list – we’d like to talk with you! Because who knows, there might be a part three to this serendipitous discovery (Figure 2).

Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in