A battle between human and HIV revealed by proteomics
Published in Microbiology
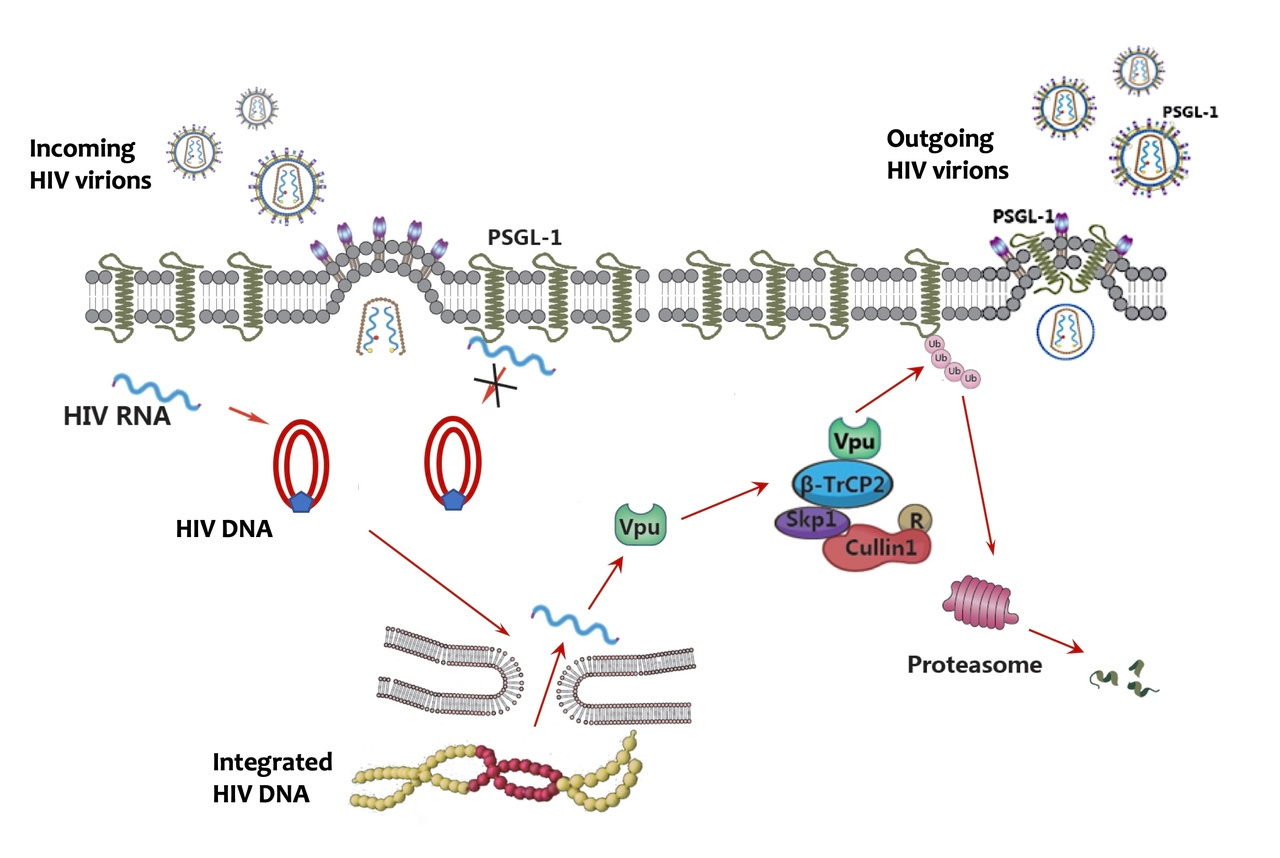
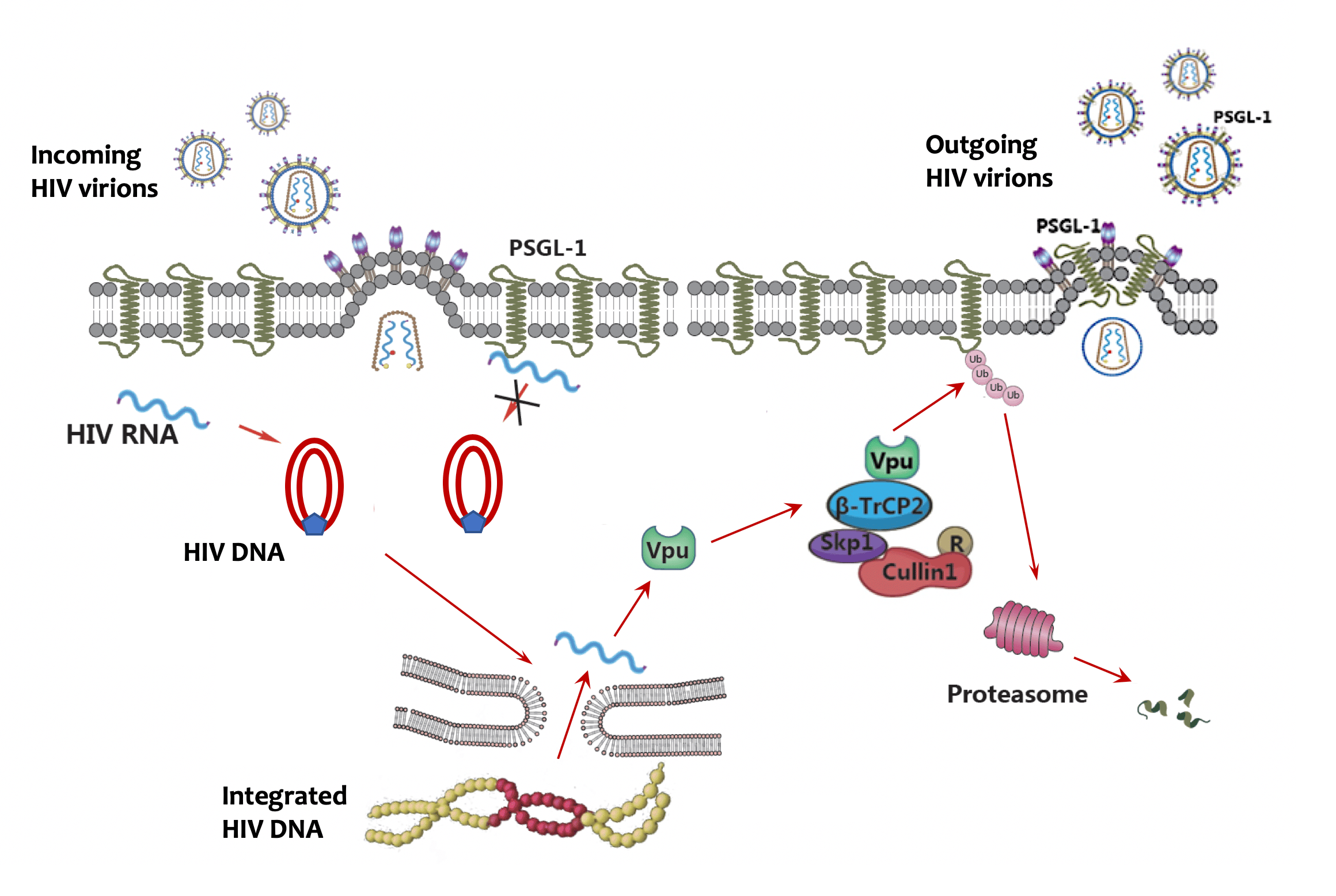
HIV infection can devastate the human immune system, which poses the question how the virus can evade the powerful immune defense. Intense research has revealed HIV as a master escape artist from the antiviral immune system. In 2002, a landmark paper from Michael Malim’s group identified an antiviral protein APOBEC3G (then called CEM15), whose function is neutralized by HIV protein Vif. This finding initiated a flurry of research to unveil APOBEC3G as a potent cytidine deaminase that can mutate nascent HIV DNA and incapacitate the virus. Unfortunately, HIV has evolved the small protein Vif to hijack the host ubiquitin-proteasome system to target APOBEC3G for ubiquitination and degradation. This APOBEC3G-Vif antagonism provides a classic example of the evolutionary arms race between virus and host because the involved genes are also rapidly evolving to gain a competitive advantage in the race. A number of HIV restriction factors have since been discovered, such as tetherin, SAMHD1 and SERINC3/5. These proteins are usually interferon induced genes and possess diverse antiviral functions. Several of them are antagonized by HIV accessory proteins, such as Vpu, Vpx or Nef. Interestingly, a common antagonizing strategy by the accessory proteins is via hijacking the ubiquitin-proteasome system to degrade the restriction factors.
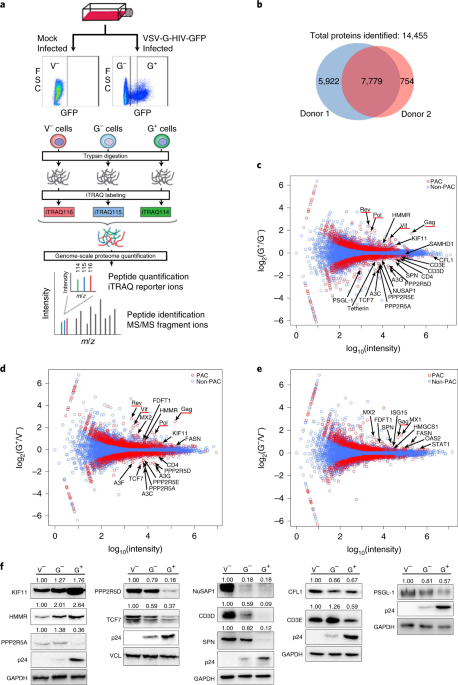
We reasoned that there might be more restriction factors degraded by HIV using a similar strategy to evade host restriction. Therefore, we applied very sensitive mass spectrometry technology to quantitate changes of the human proteome in response to HIV infection in primary CD4+ T cells, the natural host for HIV infection. This sensitive technology is ideal for the profiling of primary cells due to the limited availability of these cells. From the profiling, we identified over 14,000 proteins and found hundreds of proteins upregulated or downregulated during HIV infection, including several known restriction factors being downregulated, validating our general approach. To further narrow down the targets among the downregulated proteins, we selected those with evidence of a high evolutionary rate among primate species. Twenty-one proteins met this criterion and to our amazement, at least ten of them are known to be functionally associated with HIV, including CD4, the receptor for HIV and nine others that have been reported to have anti-HIV functions. We focused on one protein that had not been previously known to be functionally involved in HIV infection for further study. This protein, P-selectin glycoprotein ligand-1 (PSGL-1), is specifically expressed on the surface of leucocytes and mediates the anchoring of leucocytes to endothelial cells at sites of inflammation. We went on to show that HIV Vpu mediates the downregulation of PSGL-1 by hijacking the ubiquitin ligase consisting of Cul1, SKP1 and beta-TrCP2, confirming the mass spectrometry results. We also found that PSGL-1 has dual anti-HIV functions, including inhibiting viral DNA synthesis and blocking the infectivity of new virions. The latter inhibition is much more potent, probably due to the incorporation of PSGL-1 into new viral particles. This potent inhibition explains why Vpu need to degrade PSGL-1 at the later stages of the viral life cycle. Lastly, we found that PSGL-1 is specifically induced by IFN-gamma and mediates the anti-HIV function of IFN-gamma. Overall, we provide a comprehensive database demonstrating a widespread reshaping of the human proteomic landscape by HIV and identified a new HIV restriction factor with novel antiviral functions. An important lesson learned is that evolution is not only powerful in sculpting the dynamics between virus and host, but also powerful as a tool in studying virus-host interactions.
The link to the full paper:
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in