A flow-based graph neural network on biomedical network enables accurate emerging drug interaction prediction
Published in Chemistry and Pharmacy & Pharmacology

Scientific advancements and regulatory changes have led to the development of numerous emerging drugs worldwide, particularly for rare, severe, or life-threatening illnesses [1, 2]. These drugs are novel substances with unknown or unpredictable risks, as they have not been extensively regulated or used before. For instance, although hundreds of COVID-19 drugs have been developed, only six have been recommended by the FDA as of Oct 2023, such as dexamethasone and hydrocortisone. Clinical deployment of new drugs is cautious and slow, making it crucial to identify drug-drug interactions (DDIs) for emerging drugs. However, with limited clinical trial information, unexpected polypharmacy or side effects can be severe and difficult to detect.
Large biomedical networks, such as HetioNet [3], organize facts into a directed multi-relational graph, recording relationships between biomedical concepts, such as genes, diseases, and drugs. These networks can provide side information for DDIs, especially for emerging drugs. Several methods, like Decagon [4] and SumGNN [5], try to use graph neural networks (GNNs) to aggregate topological structures from the biomedical network for drug embeddings. However, without specially considering emerging drugs, their effectiveness in predicting DDIs for emerging drugs is limited.
We developed a new GNN method EmerGNN for emerging drugs’ interaction prediction using biomedical network data. Given the DDI network and biomedical network (Figure 1a), we firstly integrate the two networks to enable communication between existing and emerging drugs connected by biomedical concepts, such as proteins, diseases or other drugs, and then add inverse edges by introducing inverse types for each relation and interaction type. The two steps generate an augmented network where the drugs and biomedical entities can communicate better (Figure 1b). For a target drug pair to be predicted (for example an emerging drug u and an existing drug v), we extract all the paths between them and combine the paths to form a path-based subgraph (Figure 1c). A flow-based GNN is applied on the subgraph to trace drug features along the biomedical edges and integrate essential information along the path. An attention mechanism is applied on the subgraph edges to adjust their importance. Finally, a linear classifier predicts the interaction type between u and v based on the representations given by GNN.
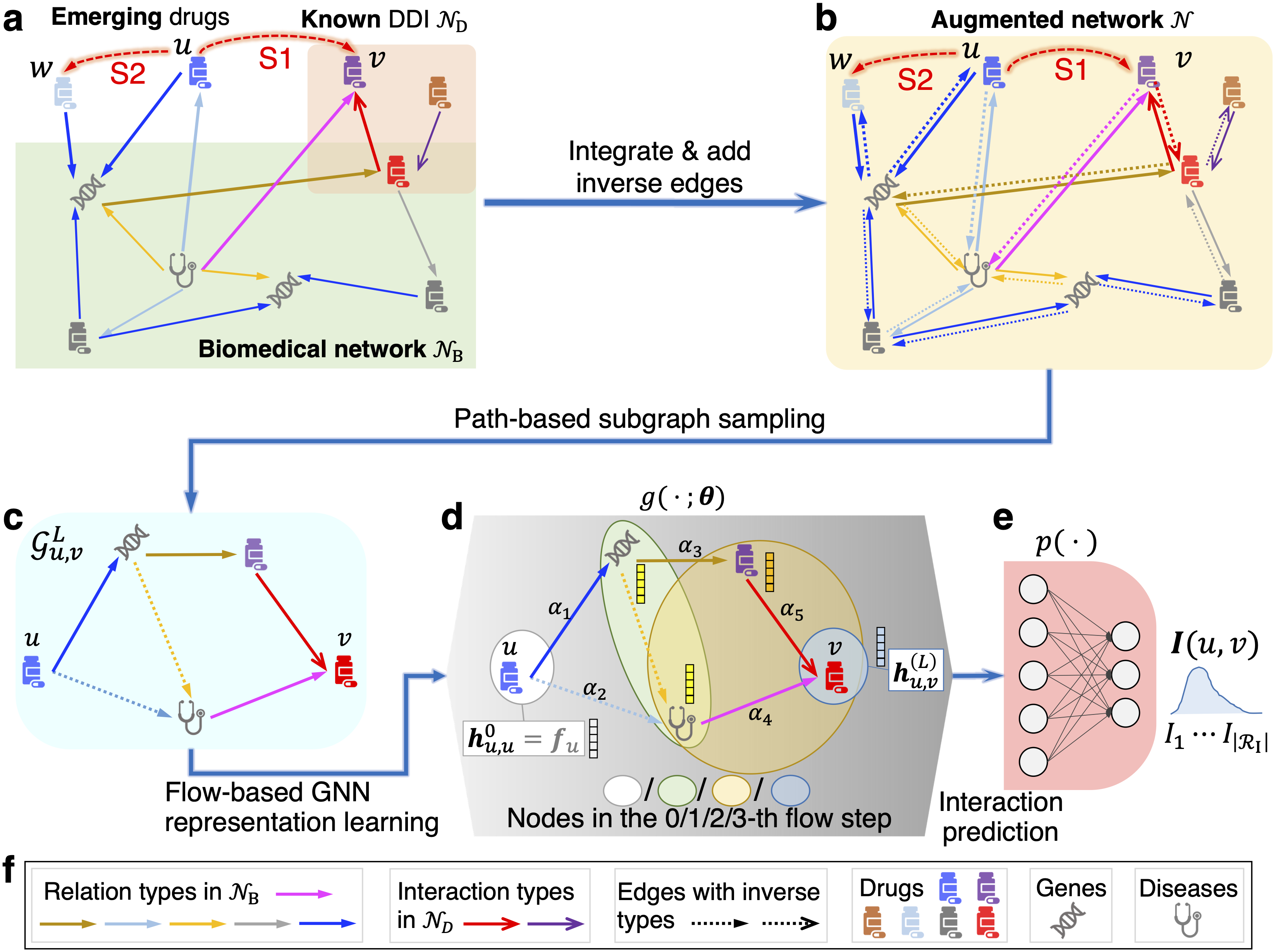
We applied EmerGNN to two DDI prediction tasks: S1 setting, determining the interaction type between an emerging drug and an existing drug, and S2 setting, determining the interaction type between two emerging drugs. EmerGNN effectively discovers the relative information from the biomedical network for emerging drug prediction. This approach achieves more accurate DDI prediction in both settings compared to existing methods, such as Decagon and SumGNN. The relevance of important paths learned by EmerGNN has been verified by correlation analysis and case studies. For example, we discovered that similarity between drugs is an important property to bring knowledge to the emerging drugs, and that the interactions “decrease the analgesic activities” and “increase the constipating activities” are highly correlated. We believe that EmerGNN’s strong prediction ability has the potential to clinically improve patient care and contribute to more efficient drug development processes.
Our study demonstrates the effectiveness of EmerGNN for predicting DDIs for emerging drugs, and suggests that graph neural networks could be a powerful tool for leveraging existing biomedical network data in low-data scenarios. One limitation of EmerGNN is that the emerging drug to be predicted should be included in the biomedical network. Building connections between emerging drug and existing drug through molecular formula or property may help address this issue. Although we demonstrate the effectiveness of EmerGNN for DDI prediction, EmerGNN is a general approach that can be applied to other biomedical applications, such as predicting protein-protein interaction, drug-target interaction and disease-gene interaction. We anticipate that the paths attended by EmerGNN can enhance the accuracy and interpretability of these predictions. We hope that our open-sourced EmerGNN can serve as a strong deep-learning tool to advance biomedicine and healthcare, by enabling practitioners to exploit the rich knowledge in existing large biomedical networks for low-data scenarios.
References:
[1] Xian Su, Haixue Wang, Nan Zhao, Tao Wang, and Yimin Cui. Trends in innovative drug development in China. Nature Reviews. Drug Discovery, 2022.
[2] Heidi Ledford. Hundreds of COVID trials could provide a deluge of new drugs. Nature, pages 25–27, 2022.
[3] Daniel Scott Himmelstein, Antoine Lizee, Christine Hessler, Leo Brueggeman, Sabrina L Chen, Dexter Hadley, Ari Green, Pouya Khankhanian, and Sergio E Baranzini. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife, 6:e26726, 2017.
[4] Marinka Zitnik, Monica Agrawal, and Jure Leskovec. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics, 34 (13):i457–i466, 2018.
[5] Yue Yu, Kexin Huang, Chao Zhang, Lucas M Glass, Jimeng Sun, and Cao Xiao. SumGNN: multi-typed drug interaction prediction via efficient knowledge graph summarization. Bioinformatics, 37(18):2988–2995, 2021.
Follow the Topic
-
Nature Computational Science

A multidisciplinary journal that focuses on the development and use of computational techniques and mathematical models, as well as their application to address complex problems across a range of scientific disciplines.
Related Collections
With Collections, you can get published faster and increase your visibility.
Physics-Informed Machine Learning
Publishing Model: Hybrid
Deadline: May 31, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in