When a cell divides, it splits exactly into two identical copies. For years, biologists have been wondering how cells accomplish this. Even in relatively simple bacteria, division has a complex nature, including elaborate protein machineries that are finely coordinated in time and space. In E. coli, division is initiated at midcell, where a contractile ring forms1. The so-called ‘Z-ring’, which in its early state comprises the filament-forming protein FtsZ, and the membrane anchors FtsA and ZipA, is always assembled with extreme accuracy at midcell2,3. The question is, how do these proteins even know where the cell center is located? Specialized protein complexes are used for proper localization, with the Min system acting as the main conductor.
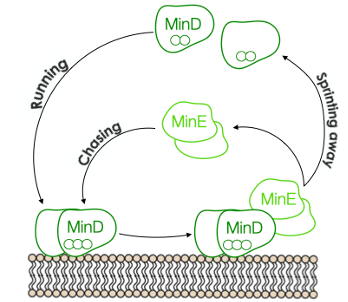
Figure 1. The MinD MinE game of catch
The Min system consists of three proteins MinD, MinE, and MinC, which oscillate back and forth between cell poles, restricting Z-rings formations at midcell4,5. The operating principle is as follows: MinD, upon dimerization, is recruited to the membrane. Once bound, MinD interacts with MinE, causing both proteins to dissociate from the membrane. The sequential interplay between these two proteins is reminiscent of a game of catch. MinE sprints MinD away from the membrane, then MinD runs out to a new location, only to be chased away again. The game between the two proteins results in oscillating gradients throughout the cell, with the highest concentrations of Min proteins at the poles and the lowest in the middle. The remaining player, MinC, tags along to the oscillating game, and its job is to prevent the formation of the Z-ring. The time-averaged concentration of MinC is the lowest at midcell, allowing the contractile ring to develop solely there6,7.
Although the essential proteins that orchestrate bacterial division are well known and many processes underlying their interconnections have been uncovered, there are still unresolved questions about how they precisely interact on the membrane to promote symmetrical scission. The rational in vitro reconstruction of Z-ring8,9components and the Min system10-12 has revealed some of the biophysical principles governing the system’s behavior. Yet, the dynamic interplay between ring-forming proteins and the Min system is poorly understood. MinC is definitely needed for correct placement of the division site in living cells13,14. Several studies, however, suggest that MinD and MinE on their own regulate the distribution of peripheral membrane proteins15-17. This made us wonder: could MinDE oscillations, alone, operate as a spatial regulator of the early ring proteins?
In our new report, we attempted to answer this question. We reconstituted in vitro the MinD and MinE oscillations, with and without MinC, in the presence of the early Z-ring components. Our study was carried out in a cell-free expression system of known composition derived mostly from E. coli. To a certain degree, this system helps mimicking the complex molecular composition of the cytoplasm. The proteins of interest were expressed in physiologically relevant concentrations using a single DNA template that combined the three genes ftsA, minD and minE. The interplay between FtsA-FtsZ and the Min system (with or without MinC) was investigated on flat membranes, also known as supported lipid bilayers, as well as inside microdroplets.
We started off the experimental section by investigating the membrane organization of FtsA-FtsZ in the presence of the whole Min system. As expected, we observed that the FtsA-FtsZ on the membrane are organized into anti-phased oscillations driven by the Min system. One explanation for this reorganization is, as previously mentioned, that the presence of the inhibitor MinC rearranges FtsZ polymers. Basically, FtsZ is restricted to the membrane area where MinC is not present, resulting in an anti-phase pattern of the Min and FtsA-FtsZ subsystems. To test the possibility that the surface waves made of MinD and MinE might be capable of spatially distributing FtsA-FtsZ on their own, we conducted the same experiment, only this time without MinC. Here too, microscopy images showed similar anti-phase dynamic patterns, suggesting that MinDE oscillations alone operate as a spatiotemporal regulator of FtsA-FtsZ (Fig. 2a). The same conclusion holds when encapsulating the two subsystems inside water-in-oil droplets (Fig. 2b). The Min and FtsA-FtsZ proteins segregate in different regions of the membrane both in the presence or absence of MinC. We then looked at how a change in FtsZ membrane anchor would affect the response to MinDE oscillations by replacing FtsA with the gain-of-function mutant FtsA* or the integral membrane protein ZipA. The results supported our previous findings, albeit the degree of responsiveness depends on the nature of the FtsZ-anchoring proteins.
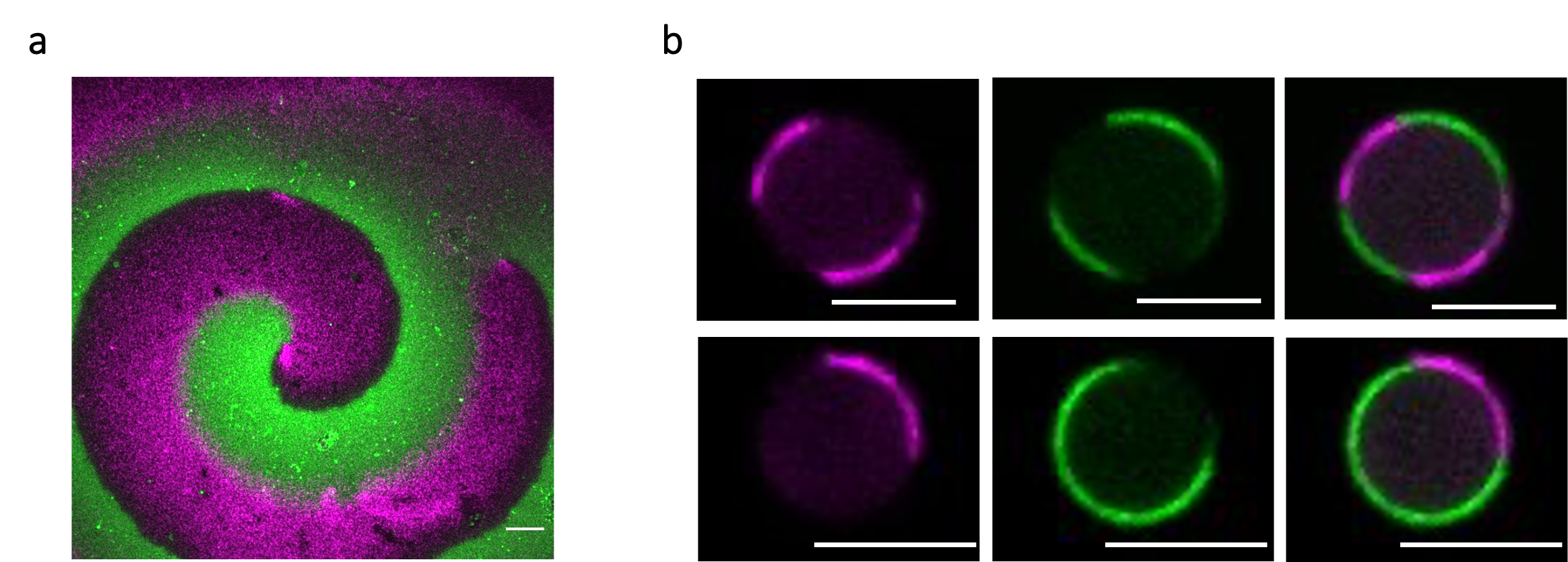
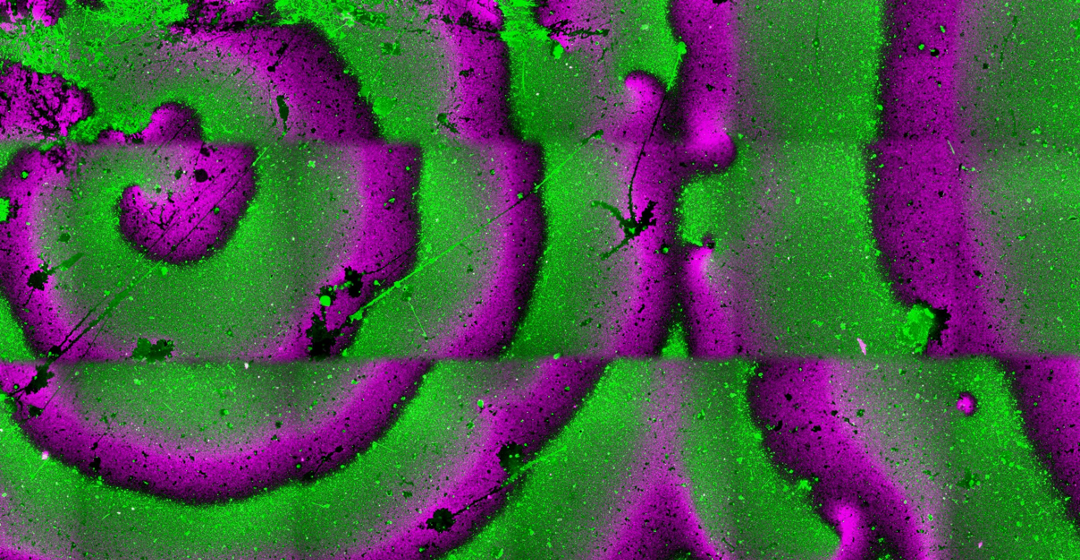
Figure 2. Fluorescence microscopy images showing antiphase spiral oscillations of FtsA-FtsZ (magenta) and MinD-MinE (green) on top of a supported lipid bilayer (a) and within two water-in-oil droplets (b). Scale bars are 10 µm.
Based on the present data, we argue that ‘the game of catch’ played by MinD and MinE has a more significant role in the spatial coordination of early division proteins, than simply distributing MinC at the cell poles (Fig. 3). Although our findings arise from in vitro reconstitution experiments, we discuss in the article the potential impact to better apprehend bacterial cell division, hoping to spark curiosity of the ‘in vivo community’ as well.
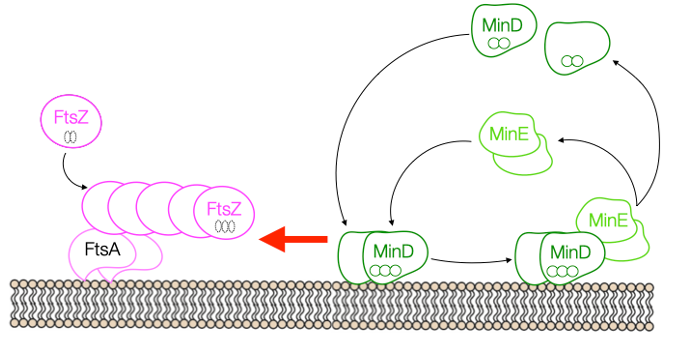
Figure 3. Our proposal: MinD MinE oscillations affect the localization of membrane-bound FtsA-FtsZ filaments
References
-
Bi, E. & Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 (1991).
-
Hale, C. A. & de Boer, P. A. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181, 167–76 (1999).
-
Ma, X., Ehrhardt, D. W. & Margolin, W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl. Acad. Sci. 93, 12998–13003 (1996).
-
Thanedar, S. & Margolin, W. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 14, 1167 (2004).
-
Rowlett, V. W. & Margolin, W. The Min system and other nucleoid-independent regulators of Z ring positioning. Front. Microbiol. 6, 478 (2015).
-
Kruse, K. A dynamic model for determining the middle of Escherichia coli. Biophys. J. 82, 618–27 (2002).
-
Raskin, D. M. & de Boer, P. A. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96, 4971–6 (1999).
-
Krupka, M., Sobrinos-Sanguino, M., Jiménez, M., Rivas, G. & Margolin, W. Escherichia coli ZipA organizes FtsZ polymers into dynamic ring-like protofilament structures. MBio 9, e01008-18 (2018).
-
Loose, M. & Mitchison, T. J. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 16, 38–46 (2014).
-
Hu, Z. & Lutkenhaus, J. Topological regulation of cell division in E. coli. spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7, 1337–43 (2001).
-
Loose, M., Fischer-Friedrich, E., Ries, J., Kruse, K. & Schwille, P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 320, 789–792 (2008).
-
Loose, M., Fischer-Friedrich, E., Herold, C., Kruse, K. & Schwille, P. Min protein patterns emerge from rapid rebinding and membrane interaction of MinE. Nat. Struct. Mol. Biol. 18, 577–583 (2011).
-
De Boer, P. A. J., Crossley, R. E. & Rothfield, L. I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56, 641–649 (1989).
-
Hale, C. A., Meinhardt, H. & de Boer, P. A. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20, 1563–72 (2001).
-
Lee, H. L. et al. Quantitative proteomics analysis reveals the Min system of Escherichia coli modulates reversible protein association with the inner membrane. Mol. Cell. Proteomics 15, 1572–1583 (2016).
-
Ramm, B. et al. The MinDE system is a generic spatial cue for membrane protein distribution in vitro. Nat. Commun. 9, 3942 (2018).
-
Shih, Y. L. et al. Active transport of membrane components by self-organization of the Min proteins. Biophys. J. 116, 1469–1482 (2019).
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in