A journey into the fascinating world of polar diatoms
Published in Ecology & Evolution

Our knowledge about the ecology and evolution of marine microbes is dominated by data from species that live in temperate and tropical oceans. To work with polar microbes requires significant logistic efforts to study them in their natural environment, and it also requires specific equipment to work with them in the laboratory. I was fortunate enough to conduct the work for my PhD thesis at the Alfred-Wegener Institute for Polar and Marine Research (Bremerhaven, Germany) where both logistics for field work and the most appropriate equipment was available to explore the adaptation and evolution of polar diatoms in the laboratory. Before I began to work on my thesis, my interest in polar marine ecosystems had been spurred by several expeditions to the Arctic Ocean where I worked on the ecology and physiology of sea-ice algae. Sea ice is a fascinating habitat because it is a semisolid matrix created from liquid sea water, which will end up being liquid sea water again when it melts. Not many species globally are exposed to similar habitat transformations under extreme environmental conditions, including strong seasonal fluctuations in light, temperature, salinity, essential gases (e.g. oxygen and carbon dioxide) and nutrients (e.g. nitrate, iron). Although many of us might consider polar sea ice as an exotic habitat, at its maximum it covers 13% of the Earth surface (similar to the size occupied by tropical rain forests or deserts), making it one of the major biomes on the planet. However, its extent is under threat due to global warming (Fig. 1).
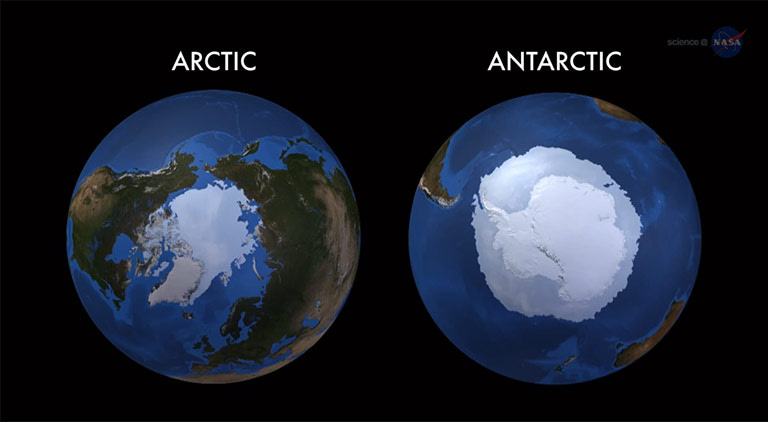
Figure 1. In 2014, Antarctic sea ice reached a record maximum extent while the Arctic reached a minimum extent in the top ten lowest since satellite records began. Credit: NASA's Goddard Space Flight Center.
The main primary producers in polar oceans are diatoms, which in general can be considered as cold-loving organisms as they thrive wherever there is turbulent, cold and nutrient-rich water. Thus, polar oceans are their natural home where they underpin most of the food webs (aquatic and terrestrial), as terrestrial primary production in polar regions does not provide enough food for these complex and productive ecosystems. As the oceanographer Alfred Bigelow once said "all fish are diatoms”, because diatom blooms fuel the growth of herbivorous zooplankton and they in turn are prey for larvae of invertebrates (e.g. krill) and fish. How diatoms have evolved to cope with polar marine conditions and especially the extreme habitat transformation between sea water and sea ice was largely unknown.
My interest in beginning a genome project with a diatom species that was known to be a key player in polar oceans and an obligate cold-loving species (psychrophile) began about 2000 when the first complete genome sequences were published.
I recognized the opportunities but was less aware of the challenges in terms of undertaking a genome project with a marine organism at that time. In 2005, I arrived with my wife Claudia and our 2-year old son Paul as a young PostDoc in the laboratory of Ginger Armbrust (University of Washington, US) who at that time had just published the first diatom genome sequence (Thalassiosira pseudonana) [1]. In 2007, we received a sequencing grant from the Joint Genome Institute (JGI, US Department of Energy's Office of Science) to sequence the genome of Fragilariopsis cylindrus, the first psychrophilic eukaryote to be sequenced (Fig. 2).

Figure 2. Scanning electron micrograph of two cells of Fragilariopsis cylindrus. Image courtesy of Gerhard Dieckmann.
JGI under the leadership of Igor Grigoriev applied Sanger sequencing to provide a first draft assembly (V1) http://genome.jgi.doe.gov/Fracy1/Fracy1.home.html that became available for analyses by 2008. At that time, my family and I were just moving to the University of East Anglia (UK) where I started to lead my own laboratory in the School of Environmental Sciences (ENV). When my colleagues and I started to annotate the genome of F. cylindrus, we were excited to find genes and gene families that were not present in any of the diatom genomes that had been sequenced before. I still remember the discovery of a proton pumping rhodopsin in this genome, which had never been identified in any photosynthetic marine alga. This discovery led to a PhD studentship for Jan Strauss and several international collaborations to characterize the function and significance of this protein for marine phytoplankton. We also discovered many different ice-binding and antifreeze proteins, which help this species and other polar algae to survive the harsh conditions in sea ice. However, it took the careful look of Beverley Green (University of British Columbia, Canada) at the group of genes encoding light harvesting proteins to find out that this genome was very different from anything that had been sequenced before, at least in the marine environment. Beverley found that this group of genes was greatly expanded but with the help of JGI we found that some of these paralogs were in fact diverged alleles of the same gene. When Jeremy Schmutz and his colleagues at JGI assembled the genome, they were unable to collapse a significant number of scaffolds corresponding to both haplotypes of this diploid genome. This led to the realization that the difficulty was due to the presence of large numbers of diverged alleles that stumped the assembly programs.
In 2011, Cock van Oosterhout and his team of evolutionary geneticists came to ENV. Over the following years, they helped us to understand how the diverged alleles in the genome of F. cylindrus evolved. Almost a quarter of the genome contains alleles with sequence divergences of up to 6%. These alleles were differentially expressed across conditions that are characteristic of sea ice and polar marine ecosystems in general, such as freezing temperatures and darkness. We also found that those conditions were causing the allelic differentiation, and as the effective population size of this diatom was estimated to be huge, we concluded that there must be an allele for every occasion. This might be the reason why F. cylindrus has been so successful in such an extreme ecosystem, in which conditions change over a seasonal cycle and have continued to change over geological time.
Because these evolutionary processes have never been observed in a marine organism before, and because they had been only rarely observed in other organisms, we needed to re-sequence the genome of F. cylindrus using a sequencing technology (PacBio) that can assign allelic variants to separate chromosomes to confirm the existence of the diverged haplotypes. This work was funded and carried out by the Earlham Institute in Norwich (UK) in the group of Matt Clark under the leadership of Pirita Paajanen.
With these data, we were able to publish our paper [2] after a long journey I dared to embark on as a young PhD student in Germany about 16 years ago. It required living in three different countries, to obtain funding from very different sources and the enthusiasm, patience and persistence of my co-authors and my family to be able to publish this paper. Finding that haplotyes in diploid organisms are not redundant entities but essential for the survival and evolution of species in variable environments probably will have an impact on how we study organisms and analyse their genomes, and also how we assess their potential to cope with global change.
References
1. Armbrust, E.V., et al. (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution and metabolism. Science, 306, 79-86.
2. Mock, T., et al. (2017) Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature, doi:10.1038/nature20803 http://www.nature.com/nature/journal/vaop/ncurrent/full/nature20803.html
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.


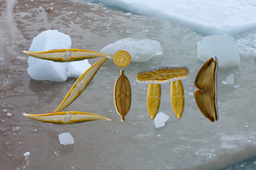
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in