A search for anti-CRISPRs
Published in Microbiology
The paper in Nature Microbiology is here: http://go.nature.com/2wNCOxg
There’s a running joke in the Moineau lab that a postdoc is someone who finds a dozen of reasons why something will fail, but winds up trying it anyway. Whether the whole thing is funny or not depends largely on the outcome of your experiments that day.
When I started in Sylvain Moineau’s group at Université Laval (Québec city, Canada), there was already buzz in the CRISPR field about the notion of anti-CRISPRs: molecules able to shut off the activity of the now widely used genome editing tool, Cas9. Bacterial viruses (phages) would be the natural place to look for them – CRISPR-Cas systems are, after all, largely an anti-phage immune system. Still, I remember being quite confident that our search would fail. Why would phages, which can easily bypass CRISPR-based immunity with a single point mutation, waste precious genomic information encoding an off-switch? It wasn’t long before our first tentative forays into this field were justified by the landmark discovery of phage-borne anti-CRISPRs in the Davidson & Maxwell Labs at the University of Toronto (Canada).
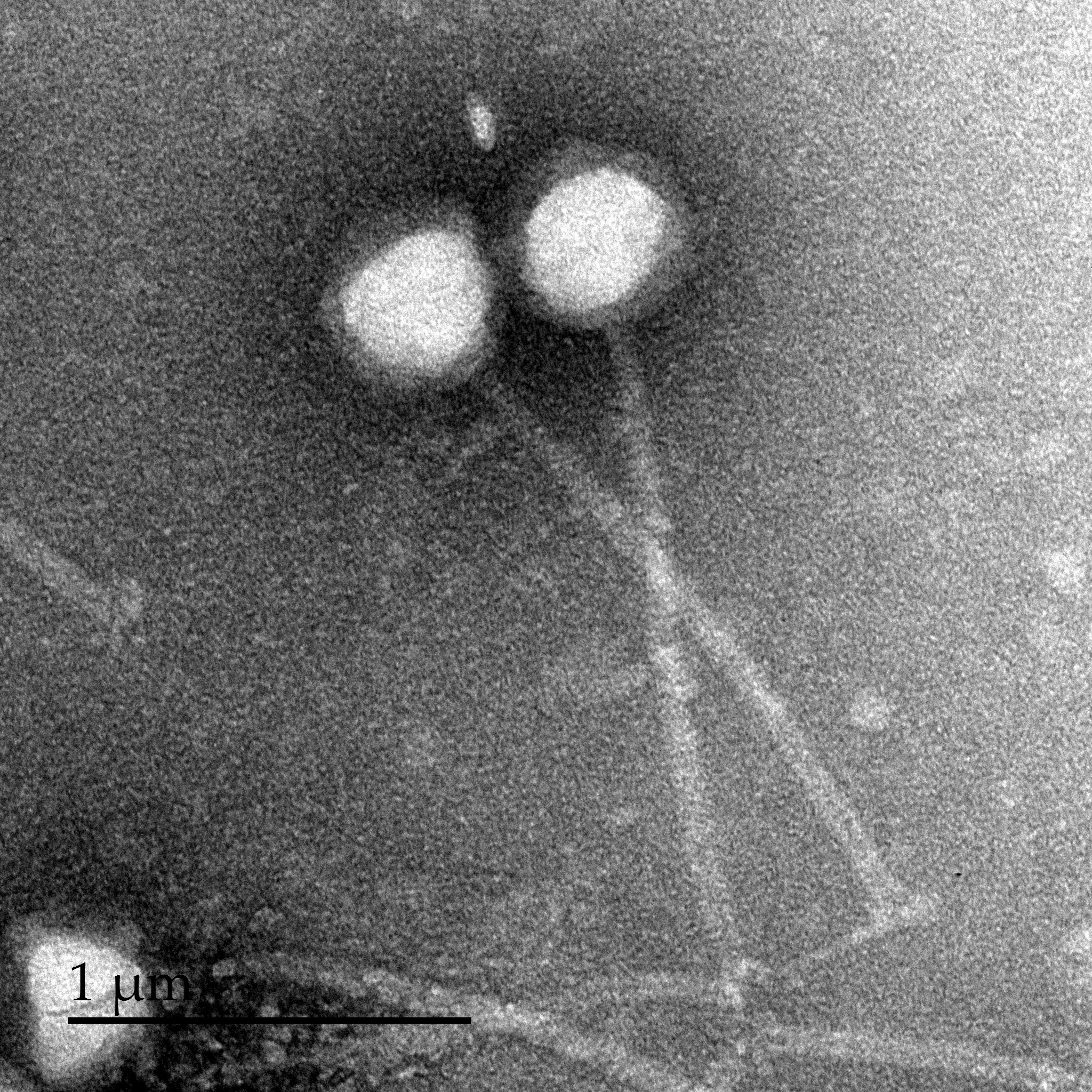
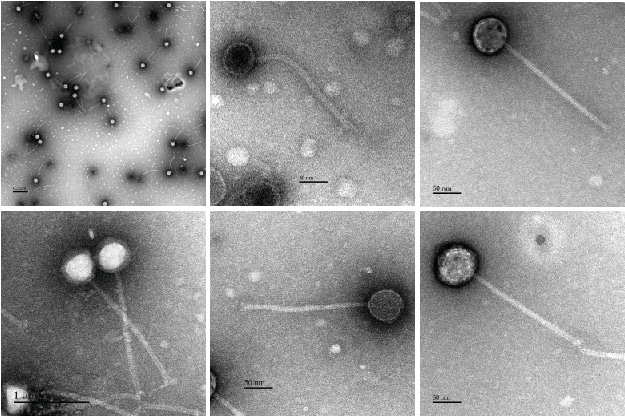
Our own search intensified, and thanks to a long-standing (>15 years) and very successful collaboration of the Moineau lab with DuPont™ France and USA (previously Rhodia and Danisco), we had access to unique phages infecting Streptococcus thermophilus strains. Of interest were sets of virulent phages infecting the same S. thermophilus strain, but resulting in different CRISPR-based survival rates in the host; some property of the phage was influencing the host’s ability to gain resistance to it using CRISPR. The postdoc strikes again; I had dozens of (excellent!) reasons why this wouldn’t be due to anti-CRISPRs… but I had to be sure. We developed a means to CRISPR-immunize the strain in question against a full set of phages, and see if some phages could overcome the pre-existing immunity. One did.

Obviously, we had to find out what phage component was responsible for the anti-CRISPR function – and again, I had a laundry-list of reasons why we wouldn’t: it could be encoded for by multiple loci, toxic to the host, non-CRISPR specific (e.g. a large-scale transcriptional repressor), etc. We systematically cloned and expressed several phage genes. We wound up answering a question we hadn’t set out to ask: how many of our phage’s genes can be stably expressed in its host? (86%, and climbing). And, as luck would have it, we found our anti-CRISPR: AcrIIA5.
The story of ‘certain’ failures that turned out otherwise doesn’t end there; when we found that its activity against one of the two active CRISPR-Cas systems carried by S. thermophilus was very weak, we had every reason to believe it wouldn’t inhibit the closely related genome-editing Cas9 of Streptococcus pyogenes – but it did.
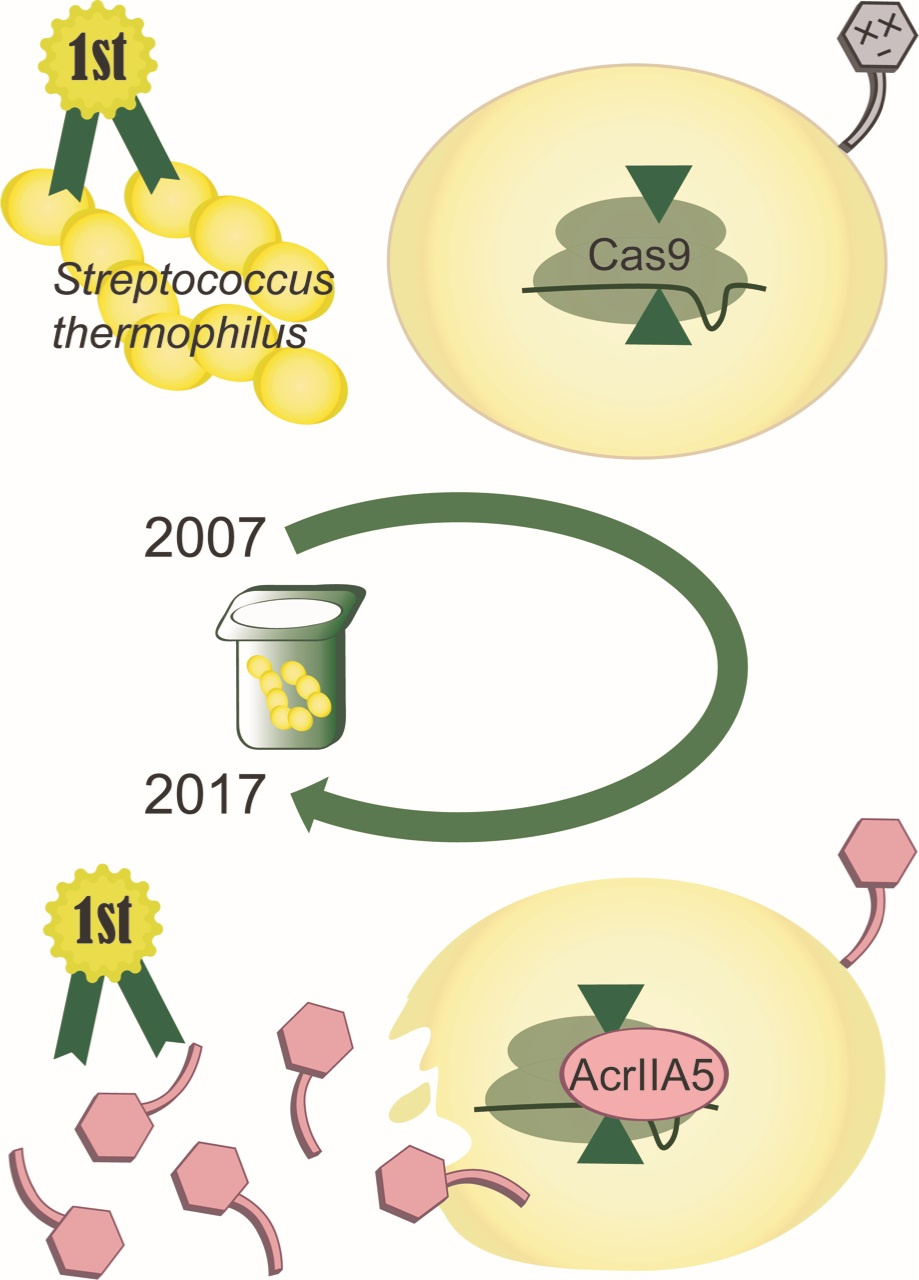
Don’t let the story mislead you though, the overwhelming majority of experiments I have been convinced would fail did. But when being wrong would be interesting enough, we eagerly try anyway, despite the odds. And while we’ve made great strides, identifying an anti-CRISPRs capable of regulating the activity of the S. pyogenes Cas9, and the first anti-CRISPR of its kind, I’m still looking back at my initial reaction to anti-CRISPRs. I was wrong about them existing – and glad to be wrong! – but I still don’t really know why they exist…
The paper discussed is available here.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in