A single prokaryotic E2 super enzyme imitates the entire ubiquitin pathway to regulate the antiviral cGAS
Published in Microbiology, Cell & Molecular Biology, and Immunology
The cGAS-STING pathway sensing infection and activating innate immune responses in mammals was evolved from a bacterial CBASS defense system. CBASS relies on cGAS/DncV nucleotide transferases (CD-NTases), which produce cyclic oligonucleotides as antiviral signaling in response to phage infection to activate an effector, and enable it to perform the killing function by degrading phage and host DNA, destroying cell membrane integrity, or consuming the essential molecule NAD+, thus providing immunity to the bacteriophage through abortive infection. The activity of CD-NTases enzymes is tightly regulated within the cell, and most CBASS operons encode accessory proteins in addition to the CD-NTases and effectors. Some of these ancillary components have been reported to function as a threat sensor or a regulator of bacterial defenses; however, most of their functions and mechanisms are unclear. CBASS systems have been identified in 14% of sequenced bacterial and archaeal genomes, analysis of diversity showed that 58% of them encoded accessory proteins, which usually contain domains homologous to eukaryotic proteins. Among them, 67% CBASSs encode homologs of the ubiquitin system.
The ubiquitin system is a typical protein post-translational modification system in eukaryotes and one of the important features that distinguish eukaryotes from prokaryotes. Modification of proteins by Ub/Ubls has been identified in all known eukaryotes, and they are involved in multiple processes such as protein degradation, gene expression, transcriptional regulation, and protein transport. The mechanisms of ubiquitination and deubiquitination are highly conserved, and all Ub/Ubls known to date are covalently linked to the substrates through related enzymatic pathways. The ubiquitination pathway contains a three-step cascade: Ub/Ubls are mostly synthesized as inactive precursors that need to be processed at their C-termini to expose the glycine carboxylate that is the site of substrate conjugation, which is performed by deubiquitinating enzymes or Ubl specific proteases; This site is adenylated by E1 with ATP to form a high-energy Ub/Ubl-AMP intermediate which is quickly attacked and covalently bound by the catalytic cysteine of the E1, creating a thioester linkage; The Ub/Ubl is transferred to the catalytic cysteine of the E2 via a transthiolation reaction; The Ub/Ubl can then be ligated to a substrate with the aid of the E3, resulting in the covalent isopeptide linkage of the Ub/Ubl’s C-terminus to the ε-amino group of a lysine in the substrate.
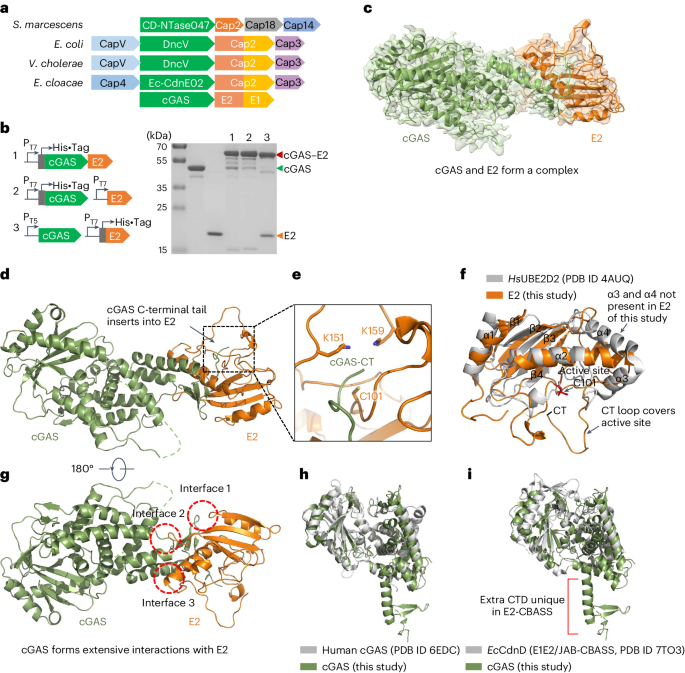
CBASS systems encoding domains homologous to the eukaryotic ubiquitin machinery can be divided into two main classes: one encodes a Cap 2 that comprises a fusion between E1 and E2 domains, and a Cap 3 that contains a protein domain predicted as an isopeptidase of the JAB/JAMM family (E1E2/JAB-CBASS); the other only encodes a Cap 2 protein contains an E2 domain (E2-CBASS). For the former, previous studies have shown that Cap2 activates the cGAMP signal molecule synthesis by conjugating cGAS to other proteins, while Cap3 antagonizes the activity of Cap2 by dissociating cGAS; For the latter, its function is elusive.
In the published paper “Phage defense system CBASS is regulated by a prokaryotic E2 enzyme that imitates the ubiquitin pathway”, we sought to elucidate the role of the CBASS-related E2 protein.
Interestingly, when studying the interaction between cGAS and E2, they were found to form a covalent complex. The structure of the cGAS-E2 complex showed that the C-terminus of cGAS is threaded into a pocket of E2. The two regions have a conserved glycine and cysteine, respectively, which subsequently proved to play a key role in the covalent linkage between cGAS and E2. However, the covalent bond formed between cGAS and E2 is not a thioester bond, and the glycine was removed during the covalent linkage, which quite surprised us. We further found that the covalent bond formed between cGAS and E2 is actually an isopeptide bond, and that the conserved glycine and cysteine function as recognition and catalytic sites, respectively, which is obviously different from the eukaryotic ubiquitin system and E1E2/JAB-CBASS system, and rather more similar to the protein ligation mechanism catalyzed by the bacterial sortase.
Furthermore, we demonstrate that cleavage of the carboxyl-terminal residues of cGAS during the covalent linkage is catalyzed by the cysteine protease activity of E2, and that the active sites are only conserved in E2 from the E2-CBASSs, but not in the known eukaryotic E2 and E2 from the E1E2/JAB-CBASSs. We also found that mutation of the conjugation site lysine of E2 triggers the formation of poly-cGAS, similar to the formation of eukaryotic polyubiquitin chains. Thus, we demonstrated that the bacterial E2 in our study regulates cGAS by imitating the ubiquitination cascade, including the cleavage of the C-terminus of cGAS or the isopeptide bond (functions like a protease), conjugation of cGAS to a cysteine residue (functions like E1 and E2), ligation of cGAS to a lysine residue (functions like E3 and a target protein), and poly-cGASylation (Fig. 1). This multifunctional E2 provides compelling evidence for the central role of E2 in ubiquitin systems, and provides insights into the evolution of eukaryotic ubiquitin system.
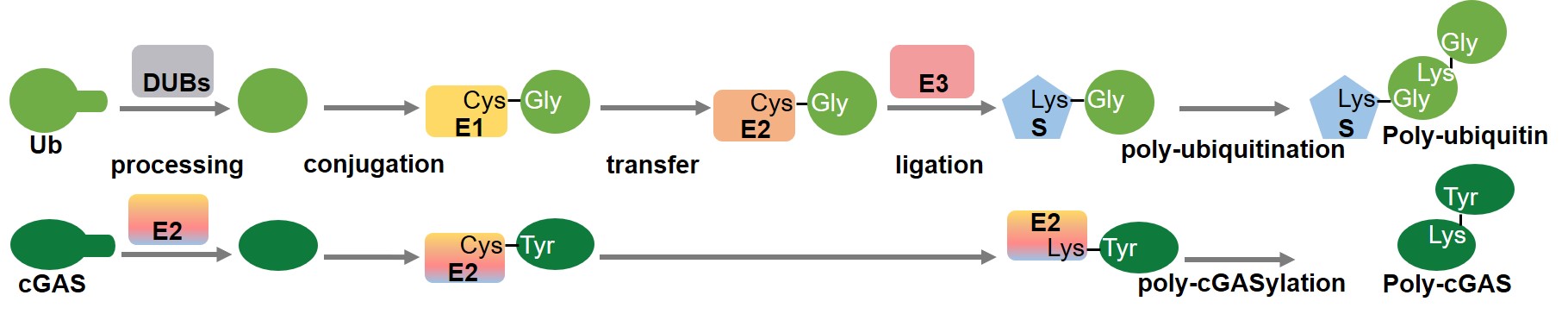
Fig. 1 Schematic diagrams showing the eukaryotic ubiquitin pathway (top) and E2-mediated cGAS regulation (bottom).
Our work also reveals a defense mechanism by which the CBASS system encoding the E2 protein protects bacteria from phage infection. Upon phage infection, E2 mediates the formation of poly-cGAS, which significantly activates the synthase activity of cGAS, the sequential cGAMP production triggers membrane disruption and abortive infection (Fig. 2). Compared to the reported mechanism of E1E2/JAB-CBASS, we reveal that E2-CBASS in this work utilizes a more concise and specific mechanism, with distinct protein chemistry, to regulate the anti-phage signaling.
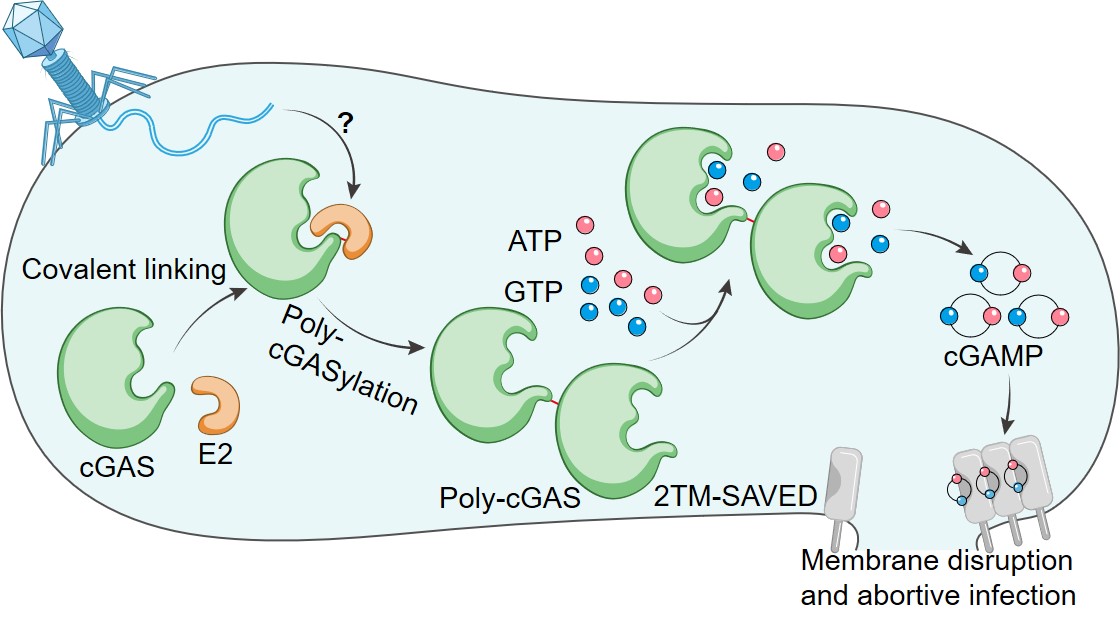
Fig. 2 Model for the E2-CBASS antiviral mechanism.
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in