A versatile CRISPR platform enables tailored genetic screens in vivo to identify novel virulence factors in host-pathogen interaction
Published in Microbiology

Written by Joanna Young and Moritz Treeck
Toxoplasma gondii is one of the most successful parasites on earth, infecting up to 1/3 of the human population. While in the majority of infections the parasite causes mild symptoms and is readily controlled by the host immune system, it can cause severe disease in the immune-compromised, the unborn child and when people get infected with hyper-virulent strains. In addition to being a human pathogen, Toxoplasma can infect virtually any warm-blooded animal although sexual reproduction can only occur in felines.
The ability of Toxoplasma to infect a wide range of animals represents a substantial reservoir of the parasite in the wild. The flip side is that Toxoplasma needs to be equipped to survive the attack by all different immune-defence mechanisms of the species it encounters. It does so by injecting proteins into the host cell, interfering with numerous host cell processes such as transcriptional response to the infection, cell cycle and motile behaviour of the infected cell. These parasite proteins that help facilitate successful infection are called virulence factors.
Toxoplasma expresses 150-300 potential virulence factors that are thought to be exported or injected into the host-cell and are likely to have a function in how the parasite interacts with the host. But how do we find the genes important for the infection of a mouse or a human? How does do Toxoplasma virulence factors help the parasite distinguish between infection of an immune-cell compared to a neuron? And which proteins make one parasite strain more virulent than another?
Genetic mapping of regions associated with virulence has proven to be a powerful tool in the past, although this won’t allow us to identify factors required by all strains. Additionally, targeted, hypothesis-driven gene deletion and subsequent testing of each of the knock-out parasite strains in mice has confirmed the vital roles of some secreted proteins. However, in many instances this has failed, leading to wasted time, and importantly, wasted research animals.
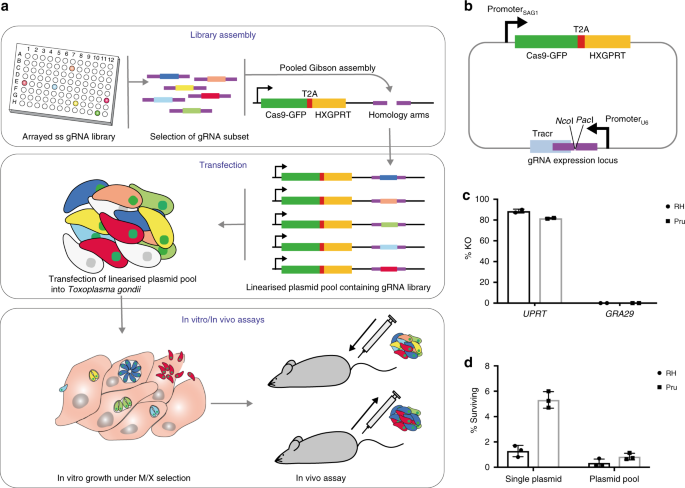
To be able to test many potential virulence factors at the same time, we sought to perform a targeted in vivo CRIPSR screen. To do this we designed a fast, flexible and cost-effective way to generate pools of mutant Toxoplasma parasites using single-stranded oligos arrayed out in multiwell plates for Gibson assembly of the mutant library KO vectors. The vector pools are transfected into parasites to generate pools of mutant Toxoplasma parasites. We first assessed the mutant viability in cell culture to deplete the pool of those mutants unable to grow under these conditions. After this first selection step the remaining mutant pool was injected into mice and parasites were retrieved after 5 days of growth. By sequencing the mutant pool before and after growth in cell culture and mice, we were able to identify those genes important for the interaction with the host.
Our strategy provides a very powerful tool for further in vivo CRIPSR screens and targeted screens in cell culture and also a major step forwards in the reduction of research animals. Pools of mutants can now be analysed together and monitored over time and in various combinations, which should prove a useful strategy to identify virulence factors in many pathogens.
One very interesting observation we made is that some mutants, previously shown to demonstrate a clear virulence defect when injected as homogenous populations appeared to survive normally when growing in mice as part of a pool. Therefore, some proteins are important for the survival within the infected cell, while others may be more important to set the overall conditions in which Toxoplasma can grow within the infect host. This was surprising and true for several members of a protein complex that contains MYR1, an important protein required for the export of a large proportion of virulence factors into the host cell. More work needs to be done to understand this phenomenon, but it is a truly exciting starting point to tease apart the functions of these virulence factors in the context of a more complex infection. This is exciting from an evolutionary angle as well as growth of less fit parasites appears to be supported by the population. More generally, the phenotype loss of individual mutants in a pool of cells may be a phenomenon observed in other CRISPR screens and may mask phenotypes previously observed for homogeneous KO populations.
The ability to identify individual mutant parasites by their gene targeting guide RNA as a molecular barcode now allows us to perform single cell sequencing and analyse the effects of a single parasite mutant within the cell it has infected. In the future, we will have the ability to tease apart the transcriptome of the host cell as an individual relationship with hundreds of different Toxoplasma mutants in various Toxoplasma strains. This unprecedented analysis power will open a window to understanding the functional relationship of Toxoplasma virulence factors with a multitude of host cell organisms, host cell types and Toxoplasma strains of clinical relevance.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in