A whole-body micro-CT scan library captures the skeletal diversity of Lake Malawi cichlid fishes
Published in Ecology & Evolution and Zoology & Veterinary Science

Lake Malawi cichlids are exceptionally diverse, ranging dramatically in body shape, size, morphology, colouration and behaviour. In just 800 thousand years, an estimated 850+ species have diverged from a single common ancestor. Remarkably, this diversity has arisen in a background of very low genetic variation, just a fraction of a per cent, a range comparable to that within human populations! I must admit, however, and perhaps to my shame, to being completely unaware of their existence before starting my PhD with Dr Berta Verd (University of Oxford). Both my undergraduate and master’s degrees are in biochemistry – but during my studies I became disillusioned by what I perceived to be excessive reductionism. I started my PhD programme at the University of Oxford, only knowing I wanted to do something in evolutionary developmental biology after I had become fascinated by the integrative understanding greatly valued by the field as an undergraduate. Luckily, I chose to do a short research project characterising Hox gene sequences in cichlids and I instantly fell in love with them; I’ve not looked back.
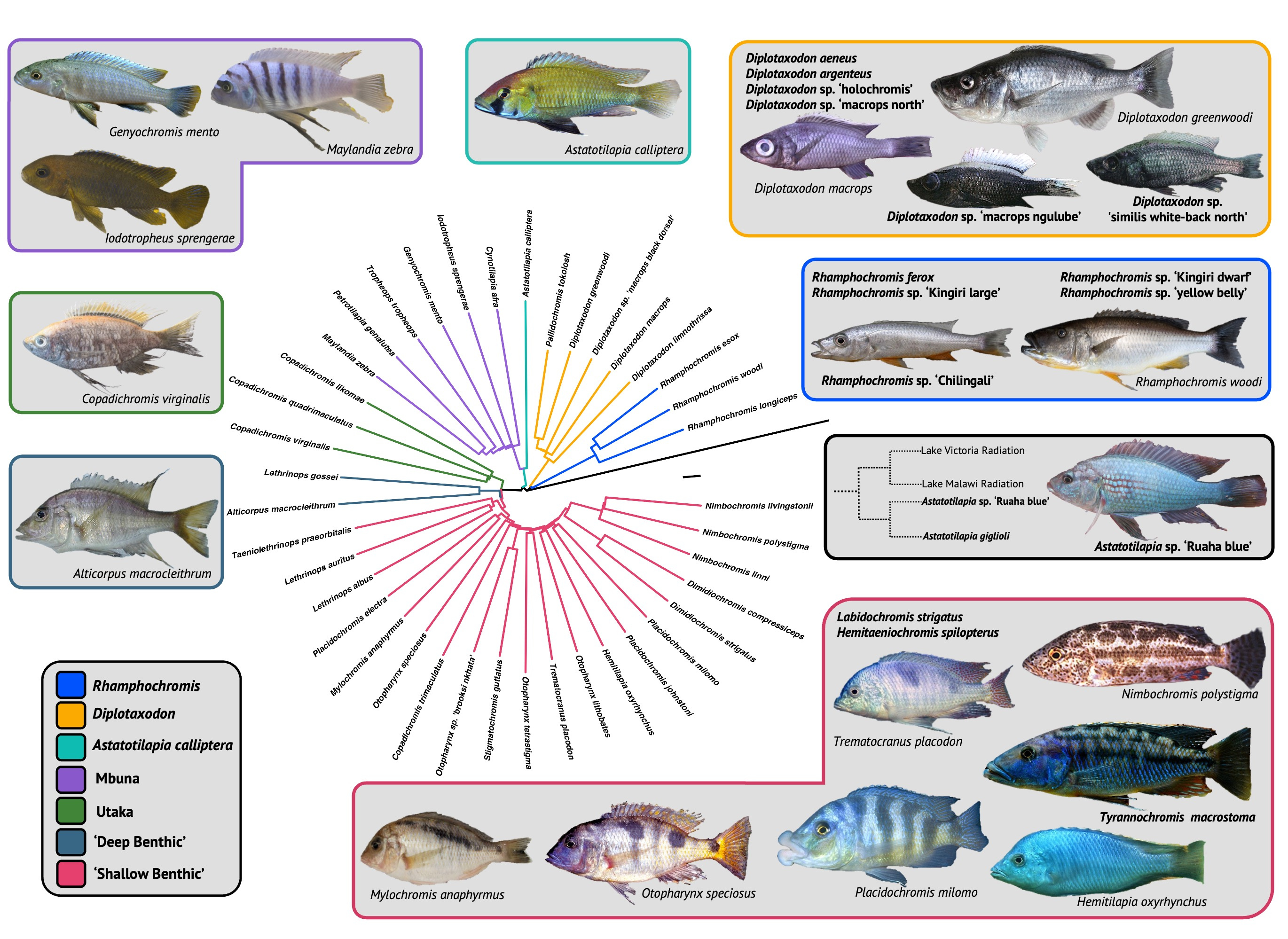
Our paper describing a dataset of whole-body µCT-scans of 116 individuals (56 species, 26 genera) of Lake Malawi cichlid was recently published in Scientific Data. We sampled species from all seven ecomorphological groups within the radiation, sampling 7 genera of rock-dwelling, predominately algae-grazing 'mbuna', including the highly specialised scale-eating Genyochromis mento (which you can download here). We also scanned several highly specialised 'shallow benthics', such as the snail-crusher, Trematocranus placodon, and ambush predators, such as those within Nimbochromis, which partially bury themselves in the sediment luring fish with their unique colouration patterns, which mimic dead and decaying flesh. We were incredibly fortunate to also widely sample Rhamphochromis, a genus of ferocious pelagic, open-water piscivores, which boast some of the largest fish in the radiation amongst its members. We sampled both the largest and smallest members of this genus, R. woodi and R. sp. 'Kingiri dwarf', which have standard lengths of 40cm and 7.5cm, respectively, and would act as excellent models for the study of allometry, which is poorly understood, particularly in cichlids.
The processed data are freely available for anyone to download on Morphosource. These data can generate excellent quality 3D models for geometric morphometric studies of any number of bones, including the well-studied jaw and craniofacial bones, as well as bones of the under-studied axial skeleton, such as the vertebrae and pterygiophores. We have even been able to use these digital models for 3D printing. This was particularly beneficial for an undergraduate practical on the teleostean body plan, in which we printed various jaw models from the dataset to introduce the students to ecological specialisation, as well as convergent and constrained morphological evolution. Our dataset joins similar ones for the Lake Tanganyika and Lake Victoria cichlid adaptive radiations, allowing for some fascinating comparative studies, including the convergence of form in the radiations.
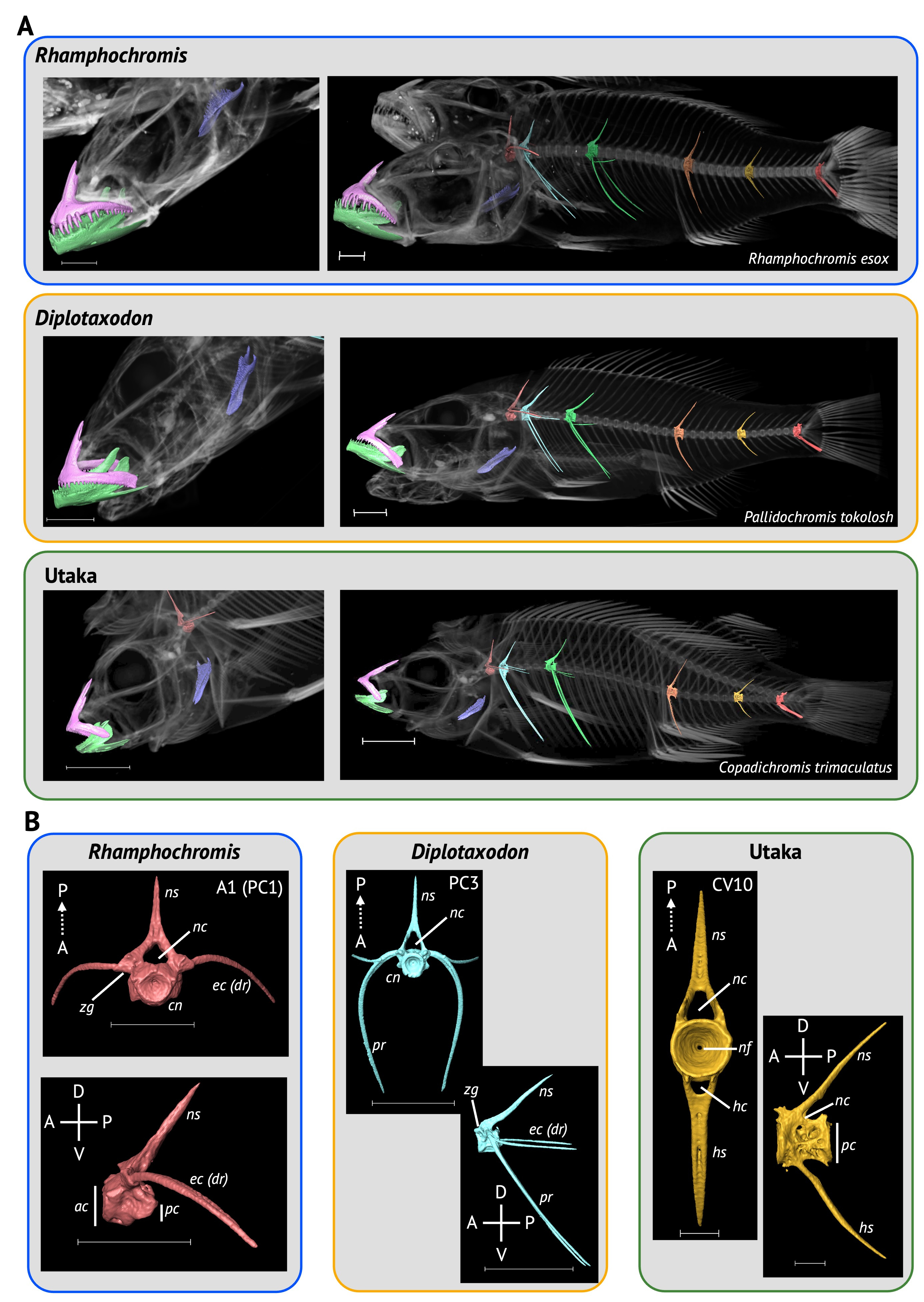
Ironically, it was not our initial intention to generate a µCT-scan dataset. We had been scanning lab-reared specimens to segment vertebrae to examine whether vertebral shape is an adaptive trait, and we haphazardly sampled species across the radiation in hopes of capturing a large proportion of the variation (in hindsight, a more systematic approach would have been preferable). Only after collecting all the data did we realise we had assembled a valuable dataset. Our data collection would not have been possible without the aid of museum and private research collections. We had been using the XTM facility at the University of Bristol to scan lab-reared specimens and had drastically underestimated how much time it would take. With very short notice, Prof Martin Genner (a co-author) kindly lent 60 (51%) of his specimens to scan, which were in his lab just a few floors above the scanner! We also took advantage of the extensive collection of Lake Malawi cichlids at the NHMUK. Our oldest specimens, two Otopharynx tetrastigma are at least 130 years old, having entered the NHMUK collection in 1893! This underscores the importance of natural history museum collections and digitisation efforts, such as µCT-scanning, can continue to keep historical collections relevant to modern research.
Our paper will lay the foundation for establishing a standardised, efficient, cheap and relatively robust methodology for scanning large numbers of specimens to a high standard. Finally, and perhaps more importantly, we hope that our dataset will become a valuable resource for other cichlid researchers, evolutionary biologists, and everyone in between. These remarkable fish consume much of my time, passion, and thought. Ultimately, the generation of this dataset would not have been possible without this sense of wonder and amazement.
Follow the Topic
-
Scientific Data

A peer-reviewed, open-access journal for descriptions of datasets, and research that advances the sharing and reuse of scientific data.
Related Collections
With Collections, you can get published faster and increase your visibility.
Data for crop management
Publishing Model: Open Access
Deadline: Apr 17, 2026
Data to support drug discovery
Publishing Model: Open Access
Deadline: Apr 22, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in