Accelerated Discovery of Novel and Selective Antimicrobials using Artificial Intelligence
Published in Bioengineering & Biotechnology
The urgency of science and the subsequent argument for accelerating discovery are abundantly clear to address many global problems. One of those pressing issues is antimicrobial resistance (AMR) – a global threat to the human health and economy. Drug-resistant diseases claim 700,000 lives a year, and they are expected to rise to 10 million deaths per year by 2050. A pandemic like COVID-19 in fact accelerates the risk of antimicrobial resistance. And while the CDC considers antibiotic resistance to be one of the biggest public health challenges of our time, unfortunately very few new antibiotics are under development to replace existing ones. To fight AMR, we need new antibiotics, and we need them soon.
The existing pipeline of preclinical antibiotic drug discovery takes more than three years, and the success rate can be as low as less than one percent. This is due to the intrinsically challenging nature of the molecular design tasks, which faces the “needle in a haystack” problem. One needs to exhaustively search the astronomical number of possible molecules and find the ideal candidate that meets multiple, often competing, objectives. To find a single novel peptide with functional activity, a minimum of 100 candidates needs to be experimentally screened.
Three years back at IBM research, a team of Artificial Intelligence (AI) researchers asked the question “can AI come to the rescue for accelerating novel antimicrobial discovery?”. To design new antibiotics, the team started developing an AI framework based on deep generative models. As no discovery is complete without real validation, we teamed up with physicists and biochemists. In the paper published in Nature Biomedical Engineering, we outline the new AI system and its validation on discovering broad-spectrum, low-toxicity, and low-resistance antimicrobial peptides (AMPs).
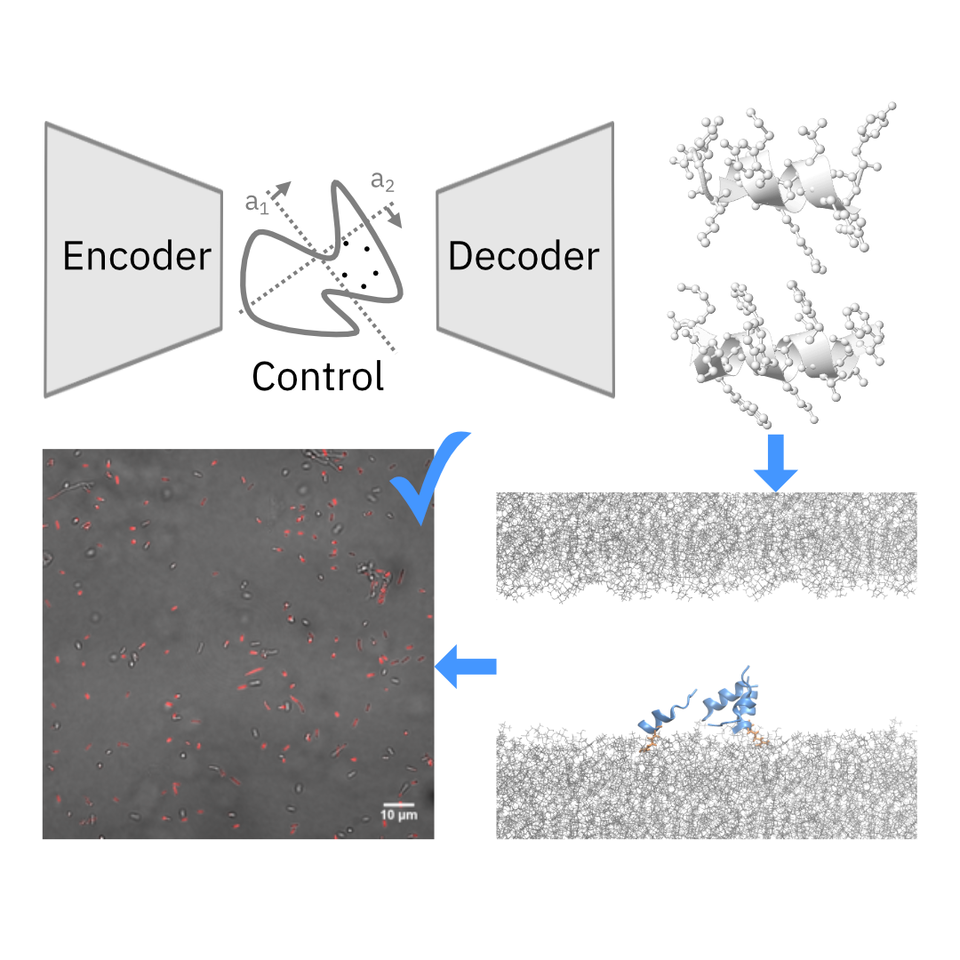
A data-driven approach to discover new AMPs is non-trivial, as it requires learning from small, noisy and/or inconsistent, and imbalanced labels. To circumvent this, we first learned a latent representation of the vast space of known peptide molecules using a generative AI model, namely a deep generative autoencoder. The goal is to create a model that captures meaningful information, such as molecular similarity and function, about diverse peptide sequences, so that the model provides the capability to explore beyond known antimicrobial templates. To evaluate and select among the deep generative models, we seek to visual Human-AI collaboration platforms. One such example can be found here.
We then employed CLaSS – Controlled Latent attribute Space Sampling— a newly developed, efficient computational method for generating novel peptide molecules with custom properties by sampling from the informative latent space of peptides. CLaSS leverages a rejection sampling scheme that is guided by the molecular property (e.g. antimicrobial) predictor trained on a model of the latent representation. Since CLaSS performs attribute-conditioned sampling in the compressed latent space, it is a computationally efficient, scalable, and easily re-purposable.
Further, we screened the AI-generated candidate antimicrobial molecules by using deep learning classifiers for additional key attributes, such as toxicity and broad-spectrum activity, to explicitly account for their preclinical therapeutic potential. To ensure trustworthiness of our AI approach, we augmented it with an independent in silico screening that is mechanistic driven. This included performing high-throughput coarse-grained peptide-membrane binding simulations for the AI-designed AMP candidates. Interestingly, there was no prior definitive recipe available to differentiate antimicrobials from non-antimicrobials in literature. Using extensive benchmarking simulations, we found novel physicochemical features, such as mean and variance of peptide membrane contacts, which are distinctive of existing antimicrobials. We used those mechanistic features to further rank the AI-designed candidates.
Within 48 days, this approach enabled us to identify, synthesize, and experimentally test twenty AI-generated novel candidate antimicrobial peptides, of which two displayed high potency against diverse Gram-positive and Gram-negative pathogens (including multidrug-resistant K. pneumoniae) and a low propensity to induce drug resistance in E. coli. No cross-resistance was seen for either of the two newly discovered AMPs, when tested using a polymyxin-resistant strain. Live-cell confocal imaging revealed formation of membrane pores as the underlying mechanism of the bactericidal mode of action of these peptides. Both antimicrobials exhibit low toxicity, as validated in vitro as well as in mice, providing important information about the safety of these antimicrobial candidates in a complex animal model. Experimental validation of the AI-designed molecules was performed in collaboration with researchers from Institute of Bioengineering and Nanotechnology at Singapore.
In summary, this work demonstrates how a synergistic approach comprised of AI models, physics simulations, and wet lab experiments can enable fast addressing a real-world discovery problem, where data availability is low, mechanistic understanding is incomplete, and trial-and-error experimental design is costly and slow. In this exciting journey, we had several ups and downs. For example, at one point we realized that our AI models were less precise due to hidden inconsistency in data labels, so we had to rework the pipeline. Techniques like this have broader potential towards fast solving other difficult discovery challenges, such as designing COVID-19 therapeutics and better CO2-capturing materials. Toward that direction, we are developing AI tools and platforms that are data-efficient, interpretable, creative, scalable and reproducible, as a means to establish and support multidisciplinary communities of discovery.
Follow the Topic
-
Nature Biomedical Engineering

This journal aspires to become the most prominent publishing venue in biomedical engineering by bringing together the most important advances in the discipline, enhancing their visibility, and providing overviews of the state of the art in each field.
Related Collections
With Collections, you can get published faster and increase your visibility.
Biosensing
Publishing Model: Hybrid
Deadline: Mar 26, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in